Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae - PubMed (original) (raw)
Phosphatase 2A negatively regulates mitotic exit in Saccharomyces cerevisiae
Yanchang Wang et al. Mol Biol Cell. 2006 Jan.
Abstract
In budding yeast Saccharomyces cerevisiae, Cdc5 kinase is a component of mitotic exit network (MEN), which inactivates cyclin-dependent kinase (CDK) after chromosome segregation. cdc5-1 mutants arrest at telophase at the nonpermissive temperature due to the failure of CDK inactivation. To identify more negative regulators of MEN, we carried out a genetic screen for genes that are toxic to cdc5-1 mutants when overexpressed. Genes that encode the B-regulatory subunit (Cdc55) and the three catalytic subunits (Pph21, Pph22, and Pph3) of phosphatase 2A (PP2A) were isolated. In addition to cdc5-1, overexpression of CDC55, PPH21, or PPH22 is also toxic to other temperature-sensitive mutants that display defects in mitotic exit. Consistently, deletion of CDC55 partially suppresses the temperature sensitivity of these mutants. Moreover, in the presence of spindle damage, PP2A mutants display nuclear localized Cdc14, the key player in MEN pathway, indicative of MEN activation. All the evidence suggests the negative role of PP2A in mitotic exit. Finally, our genetic and biochemical data suggest that PP2A regulates the phosphorylation of Tem1, which acts at the very top of MEN pathway.
Figures
Figure 1.
(A) Overexpression of PP2A components is lethal to mutants defective in mitotic exit. Saturated cell cultures were 10-time serial diluted and then spotted onto plates containing either glucose (Glu) or galactose (Gal). The plates were incubated at 25°C for 3 d. (B) and (C) Δ_cdc55_ mutant suppresses the temperature-sensitive phenotype of some mutants in the MEN pathway. The saturated cultures of strains with indicated genotype were 10-fold serial diluted and spotted onto YPD plates and incubated at indicated temperatures for 3 d.
Figure 2.
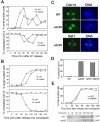
(A) Δ_cdc55_ mutant cells exit mitosis in the presence of nocodazole. Wild-type (Y300) and Δ_cdc55_ mutant (YYW28) cells in midlog phase were arrested at G1 phase with α-factor and then released into 30°C YPD medium containing 20 μg/ml nocodazole after sonication. Cells were harvested at 30-min intervals for budding index. The top panel shows the percentile of cells with extrabud and the bottom panel shows cells after 240-min incubation in the presence of nocodazole. The arrow indicates the rebudded Δ_cdc55_ mutant cell in the presence of nocodazole. (B) Top, strain 409-2-6-3 (Δ_cdc55 URA3_-tetO-112 LEU2_-tetR-GFP) was arrested at G1 phase and then released into YPD medium containing 20 μg/ml nocodazole at 30°C. The picture was taken after 240-min incubation. The spots indicate GFP marked chromosome V. Bottom, the frequency of reduplicated chromosome in wild-type and Δ_cdc55 mutants. JBY583 (_URA3_-tetO-112 _LEU2_-tetR-GFP) and 409-2-6-3 (Δ_cdc55 URA3_-teto-112 LEU2_-tetR-GFP) were arrested at G1 phase and then released into YPD medium containing 20 μg/ml nocodazole. Cells were collected every 30 min and fixed for examination of the GFP signal. (C) The rebudding phenotype of Δ_cdc55 mutants is independent of Cdc28 Y19 phosphorylation. Top, the budding index of cells with indicated genotype in YPD medium at 30°C. Bottom, the percentage of rebudded cells in the presence of 20 μg/ml nocodazole. Strains (Y300, YYW28, 397-2-2, 412-13-3) were treated as described in Figure 2A.
Figure 3.
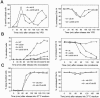
(A) Δ_cdc55_ mutant suppresses AMN1 overexpression phenotype. Strains (Y300, YFH240, YYW28, and YYW81) were transformed with either a CEN-URA3 vector or a PGAL-AMN1 plasmid. Saturated cultures of the transformants were 10-fold serial diluted and spotted onto plates containing glucose or galactose and incubated at 30°C for 2 d. (B) Mutations in catalytic subunits of PP2A result in benomyl sensitivity. Saturated cultures of strains with indicated genotype (Y300, YYW28, Y2475, and Y2762) were 10-fold serial diluted and spotted onto YPD and benomyl (15 μg/ml) plates and incubated at 25°C for 3 d. (C) Δ_pph21_ Δ_pph22_ mutants suppress AMN1 overexpression phenotype. Y300 and Y2762 were transformed with either a CEN-URA3 vector or a PGAL-AMN1 plasmid. The transformants were treated as described in Figure 3A. (D) Δ_tpd3_ mutant suppresses AMN1 overexpression phenotype. 483-15-1 (tpd3::KAN) was transformed with either a vector or a PGAL-AMN1 plasmid and the growth of the transformants was examined as described.
Figure 4.
Cdc14 is released from the nucleolus in Δ_cdc55_ mutants in the presence of nocodazole. (A**)** A1411 (CDC14-HA) and 173-1-3 (Δ_cdc55 CDC14-HA_) cells were arrested at G1 phase with α-factor and then released into YPD medium. Cells were collected every 20 min and fixed for Cdc14-HA staining. The budding index and the kinetics of Cdc14 release from the nucleolus are shown in the top and bottom panels, respectively. (B) The same strains were arrested at G1 phase and then released into YPD medium containing 20 μg/ml nocodazole at 30°C. The budding index and Cdc14 localization are shown. (C) The nuclear structure and Cdc14 localization of the 180 min samples in B are shown (top). The bottom of panel C shows the Net1 localization. G1-arrested NET1-myc and Δ_cdc55 NET1-myc_ cells were released into nocodazole medium. Cells were collected for Net1-myc staining. The nuclear structure and Net1 localization after 180-min incubation are shown. (D) Δ_cdc55_ and Δ_pph21_ Δ_pph22_ mutants fail to prevent Cdc14 from releasing in the presence of nocodazole. Midlog phase of A1411, 173-1-3, and 472-7-2 were incubated in the presence of 20 μg/ml nocodazole at 30°C. Cells were collected after 3-h incubation for Cdc14 staining. (E) Hof1 phosphorylation is more pronounced in nocodazole-treated Δ_cdc55_ mutants. G1-arrested 514-3-3 (HOF1-HA) and 514-2-3 (Δ_cdc55 HOF1-HA_) cells were released into YPD medium containing 20 μg/ml nocodazole at 30°C. Cells were collected every 30 min for protein preparation. Hof1-HA protein was examined by Western blot analysis after SDS-PAGE.
Figure 5.
CDC55 exhibits synthetic phenotype with BFA1 and BUB2. (A) G1-arrested BFA1-HA (YFH286) and Δ_cdc55 BFA1-HA_ (283-2-4) cells were released into 30°C YPD medium. Cells were taken every 20 min to prepare protein extracts. The phosphorylation status of Bfa1 were determined by Western blot analysis with anti-HA antibodies. In the bottom panel, cdc14-1 BFA1-HA cells with a vector or a PGAL_-CDC55_ plasmid were grown to midlog phase in the raffinose medium at 25°C followed by the following treatments: 1) to add galactose and incubate for 1 h at 25°C; 2) to add galactose and incubate for 1 h at 25°C and then shift to 34°C for 3 h; 3) to shift to 34°C for 3 h; and 4) to shift to 34°C for 3 h and then add galactose and incubate at 34°C for 1 h. The phosphorylation of Bfa1 protein is shown after Western blotting with anti-HA antibody. (B) Δ_cdc55_ Δ_bfa1_ and Δ_cdc55_ Δ_bub2_ double mutants display slow-growth phenotype. Cells with indicated genotype were incubated at 30°C for 2 d. (C) The localization of Cdc14 in asynchronized Δ_cdc55_ Δ_bfa1_ (443-5-1) double mutants. (D) Cell cycle-regulated Cdc14 localization in Δ_cdc55_ (173-1-3), Δ_bub2_ (221-1-1) and Δ_cdc55_ Δ_bub2_ (482-7-4) mutants. Cells were arrested at G1 phase with α-factor and then released into YPD medium at 30°C. Hydroxyurea, 200 mM, was added into the medium at 80 min when majority of the cells were large budded in order to block the release of Cdc14 from the nucleolus during the next cell cycle.
Figure 6.
Mitotic exit in Δ_cdc55_ mutants depends on MEN pathway. (A) The localization of Cdc14 in Δ_slk19_ and Δ_cdc55_ Δ_slk19_ mutants during unperturbed cell cycle progression. G1-arrested Δ464-3-2 and 450-3-1 were released into cell cycle at 30°C and 200 mM hydroxyurea was added into the medium at 80 min to block the next S phase. Cells were collected at 30 min interval for Cdc14 staining. (B) Left, the budding morphology of cells with indicated genotypes. Strains with indicated genotypes were arrested at G1 and released into YPD medium containing 20 μg/ml nocodazole at 30°C for the determination of budding index. Right, the localization of Cdc14 in the presence of nocodazole. Strains with indicated genotype were released into nocodazole medium after G1 arrest. Cells were collected every 30 min and subjected to immunofluorescence staining for Cdc14-HA. The percentage of cells with nucleolar localized Cdc14 was determined over time. (C) cdc15-2 CDC14-HA and _cdc15-2_Δ cdc55 CDC14-HA strains were arrested at G1 phase with α-factor and then released into YPD medium at 37°C. Budding index and the localization of Cdc14 were determined over time.
Figure 7.
PP2A regulates mitotic exit through Tem1 modification. (A) Δ_cdc55_ deletion partially suppresses the mitotic exit defects of tem1-3 mutants. G1-arrested tem1-3, tem1-3 Δ_cdc55, cdc15-2,_ and cdc15-2 Δ_cdc55_ mutants were released into 37°C YPD medium and the budding morphology was examined over time. The percentage of rebudded cells is shown. (B) Tem1 is a phosphoprotein. Tem1-myc fusion protein was immunoprecipitated from the cell lysate of cdc14-1 TEM1-myc incubated at 36°C for 2 h. The precipitates were incubated with λ phosphatase at 30°C for 30 min in the presence or absence of phosphatase inhibitors. Proteins were separated with 10% SDS-PAGE and then subjected to Western blotting with anti-myc antibody. (C) Δ_cdc55_ mutation leads to hyperphosphorylation of Tem1. 504-2-1 (TEM1-13myc) and 504-1-3 (Δ_cdc55 TEM1-13myc_) were arrested at G1 phase and then released into 30°C YPD medium containing 20 μg/ml nocodazole. Protein samples were prepared every 20 min with TCA method. Proteins were separated by SDS-PAGE followed by Western blot analysis. Top, the percentage of large budded cells; bottom, the modification of Tem1 protein. (D) Protein samples from 0-, 80-, and 140-min time points in C were loaded side by side and the band shift of Tem1 protein is shown after Western blot analysis.
Similar articles
- Dual Regulation of the mitotic exit network (MEN) by PP2A-Cdc55 phosphatase.
Baro B, Rodriguez-Rodriguez JA, Calabria I, Hernáez ML, Gil C, Queralt E. Baro B, et al. PLoS Genet. 2013;9(12):e1003966. doi: 10.1371/journal.pgen.1003966. Epub 2013 Dec 5. PLoS Genet. 2013. PMID: 24339788 Free PMC article. - The molecular function of the yeast polo-like kinase Cdc5 in Cdc14 release during early anaphase.
Liang F, Jin F, Liu H, Wang Y. Liang F, et al. Mol Biol Cell. 2009 Aug;20(16):3671-9. doi: 10.1091/mbc.e08-10-1049. Epub 2009 Jul 1. Mol Biol Cell. 2009. PMID: 19570916 Free PMC article. - Zds1 regulates PP2A(Cdc55) activity and Cdc14 activation during mitotic exit through its Zds_C motif.
Calabria I, Baro B, Rodriguez-Rodriguez JA, Russiñol N, Queralt E. Calabria I, et al. J Cell Sci. 2012 Jun 15;125(Pt 12):2875-84. doi: 10.1242/jcs.097865. Epub 2012 Mar 16. J Cell Sci. 2012. PMID: 22427694 Free PMC article. - Mitotic exit control: a space and time odyssey.
Segal M. Segal M. Curr Biol. 2011 Oct 25;21(20):R857-9. doi: 10.1016/j.cub.2011.09.023. Curr Biol. 2011. PMID: 22032192 Review. - Regulation of Mitotic Exit in Saccharomyces cerevisiae.
Baro B, Queralt E, Monje-Casas F. Baro B, et al. Methods Mol Biol. 2017;1505:3-17. doi: 10.1007/978-1-4939-6502-1_1. Methods Mol Biol. 2017. PMID: 27826852 Review.
Cited by
- Adaptation to the spindle checkpoint is regulated by the interplay between Cdc28/Clbs and PP2ACdc55.
Vernieri C, Chiroli E, Francia V, Gross F, Ciliberto A. Vernieri C, et al. J Cell Biol. 2013 Sep 2;202(5):765-78. doi: 10.1083/jcb.201303033. J Cell Biol. 2013. PMID: 23999167 Free PMC article. - The Polo-like kinase Cdc5 interacts with FEAR network components and Cdc14.
Rahal R, Amon A. Rahal R, et al. Cell Cycle. 2008 Oct;7(20):3262-72. doi: 10.4161/cc.7.20.6852. Epub 2008 Oct 25. Cell Cycle. 2008. PMID: 18927509 Free PMC article. - PP2A Functions during Mitosis and Cytokinesis in Yeasts.
Moyano-Rodriguez Y, Queralt E. Moyano-Rodriguez Y, et al. Int J Mol Sci. 2019 Dec 30;21(1):264. doi: 10.3390/ijms21010264. Int J Mol Sci. 2019. PMID: 31906018 Free PMC article. Review. - A new layer of regulation of chromosomal passenger complex (CPC) translocation in budding yeast.
Sherwin D, Gutierrez-Morton E, Bokros M, Haluska C, Wang Y. Sherwin D, et al. Mol Biol Cell. 2023 Sep 1;34(10):ar97. doi: 10.1091/mbc.E23-02-0063. Epub 2023 Jul 5. Mol Biol Cell. 2023. PMID: 37405742 Free PMC article. - Fin1-PP1 Helps Clear Spindle Assembly Checkpoint Protein Bub1 from Kinetochores in Anaphase.
Bokros M, Gravenmier C, Jin F, Richmond D, Wang Y. Bokros M, et al. Cell Rep. 2016 Feb 9;14(5):1074-1085. doi: 10.1016/j.celrep.2016.01.007. Epub 2016 Jan 28. Cell Rep. 2016. PMID: 26832405 Free PMC article.
References
- Alexandru, G., Uhlmann, F., Mechtler, K., Poupart, M. A., and Nasmyth, K. (2001). Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459–472. - PubMed
- Bardin, A. J., Visintin, R., and Amon, A. (2000). A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102, 21–31. - PubMed
- Charles, J. F., Jaspersen, S. L., Tinker-Kulberg, R. L., Hwang, L., Szidon, A., and Morgan, D. O. (1998). The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 8, 497–507. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous