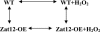The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis - PubMed (original) (raw)
The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis
Sholpan Davletova et al. Plant Physiol. 2005 Oct.
Abstract
Plant acclimation to environmental stress is controlled by a complex network of regulatory genes that compose distinct stress-response regulons. In contrast to many signaling and regulatory genes that are stress specific, the zinc-finger protein Zat12 responds to a large number of biotic and abiotic stresses. Zat12 is thought to be involved in cold and oxidative stress signaling in Arabidopsis (Arabidopsis thaliana); however, its mode of action and regulation are largely unknown. Using a fusion between the Zat12 promoter and the reporter gene luciferase, we demonstrate that Zat12 expression is activated at the transcriptional level during different abiotic stresses and in response to a wound-induced systemic signal. Using Zat12 gain- and loss-of-function lines, we assign a function for Zat12 during oxidative, osmotic, salinity, high light, and heat stresses. Transcriptional profiling of Zat12-overexpressing plants and wild-type plants subjected to H(2)O(2) stress revealed that constitutive expression of Zat12 in Arabidopsis results in the enhanced expression of oxidative- and light stress-response transcripts. Under specific growth conditions, Zat12 may therefore regulate a collection of transcripts involved in the response of Arabidopsis to high light and oxidative stress. Our results suggest that Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis.
Figures
Figure
Figure 1.
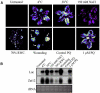
Promoter∷reporter analysis of Zat12 expression in Arabidopsis. A, Luciferase imaging in transgenic Arabidopsis plants expressing luciferase under the control of the Zat12 promoter in response to different environmental stresses. B, RNA-blot analysis of luciferase and Zat12 steady-state transcript level in the plants shown in A in response to stress. Construction of transgenic plants, stress assays, and luciferase imaging were performed as described in “Materials and Methods.” Abbreviations: Luc, luciferase; PQ, methyl viologen (paraquat); rRNA, ribosomal RNA; RWC, relative water content.
Figure 2.
Systemic induction of the Zat12 promoter in response to wounding. A, Luciferase imaging in a transgenic Arabidopsis plant expressing luciferase under the control of the Zat12 promoter in response to wounding. Arrows indicate the wounded leaves. Luciferase imaging was performed at 0, 30, and 120 min following wounding. B, RNA-blot analysis of luciferase steady-state transcript level in transgenic Arabidopsis plant expressing luciferase under the control of the Zat12 promoter in response to wounding. Luciferase transcripts are shown to accumulate in local wounded leaves (lane 2) and systemic leaves of wounded plants (lane 4). Construction of transgenic plants, wounding, and luciferase imaging were performed as described in “Materials and Methods.” Abbreviations: Luc, luciferase; rRNA, ribosomal RNA.
Figure 3.
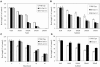
Tolerance of gain- and loss-of-function Zat12 Arabidopsis lines to abiotic stress. A, Root growth of wild-type and KO-Zat12 seedlings subjected to salinity stress. B, Root growth of wild-type and KO-Zat12 seedlings subjected to osmotic stress. C, Root growth of wild-type and KO-Zat12 seedlings subjected to heat stress. D, Root growth of wild-type and OE-Zat12 seedlings subjected to osmotic stress. Stress assays were performed as described in “Materials and Methods” using independent knockout and overexpression lines. Additional assays are shown in Supplemental Figures 1 and 2. Abbreviations: KO-Zat12-1, knockout Zat12, SALK_037357; KO-Zat12-2, knockout Zat12, SAIL 792_F04; OE-Zat12, transgenic seedlings overexpressing Zat12.
Figure 4.
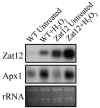
RNA blots showing the expression of Zat12 and Apx1 in H2O2-treated and untreated wild-type plants and transgenic plants overexpressing Zat12. RNA blots were performed as described in “Materials and Methods.” Abbreviations: rRNA, ribosomal RNA; WT, wild type; Zat12, transgenic plants overexpressing Zat12.
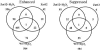
Figure 5.
Venn diagrams showing the overlap between transcript expression in wild-type plants subjected to H2O2 stress (WT + H2O2), transgenic plants overexpressing Zat12 (Zat12), and transgenic plants overexpressing Zat12 subjected to H2O2 stress (Zat12 + H2O2). Transcriptome profiling and data analysis were performed as described in “Materials and Methods.”
Similar articles
- Identification of novel transcriptional regulators of Zat12 using comprehensive yeast one-hybrid screens.
Ben Daniel BH, Cattan E, Wachtel C, Avrahami D, Glick Y, Malichy A, Gerber D, Miller G. Ben Daniel BH, et al. Physiol Plant. 2016 Aug;157(4):422-41. doi: 10.1111/ppl.12439. Epub 2016 Apr 14. Physiol Plant. 2016. PMID: 26923089 - Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis.
Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Vogel JT, et al. Plant J. 2005 Jan;41(2):195-211. doi: 10.1111/j.1365-313X.2004.02288.x. Plant J. 2005. PMID: 15634197 - Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection.
AbuQamar S, Chen X, Dhawan R, Bluhm B, Salmeron J, Lam S, Dietrich RA, Mengiste T. AbuQamar S, et al. Plant J. 2006 Oct;48(1):28-44. doi: 10.1111/j.1365-313X.2006.02849.x. Epub 2006 Aug 22. Plant J. 2006. PMID: 16925600 - The Roles of Arabidopsis C1-2i Subclass of C2H2-type Zinc-Finger Transcription Factors.
Xie M, Sun J, Gong D, Kong Y. Xie M, et al. Genes (Basel). 2019 Aug 28;10(9):653. doi: 10.3390/genes10090653. Genes (Basel). 2019. PMID: 31466344 Free PMC article. Review. - The BBX family of plant transcription factors.
Gangappa SN, Botto JF. Gangappa SN, et al. Trends Plant Sci. 2014 Jul;19(7):460-70. doi: 10.1016/j.tplants.2014.01.010. Epub 2014 Feb 24. Trends Plant Sci. 2014. PMID: 24582145 Review.
Cited by
- Global Gene-Expression Analysis to Identify Differentially Expressed Genes Critical for the Heat Stress Response in Brassica rapa.
Dong X, Yi H, Lee J, Nou IS, Han CT, Hur Y. Dong X, et al. PLoS One. 2015 Jun 23;10(6):e0130451. doi: 10.1371/journal.pone.0130451. eCollection 2015. PLoS One. 2015. PMID: 26102990 Free PMC article. - Comparative transcriptome analysis reveals the regulatory mechanisms of two tropical water lilies in response to cold stress.
Ma X, Jin Q, Wang Y, Wang X, Wang X, Yang M, Ye C, Yang Z, Xu Y. Ma X, et al. BMC Genomics. 2023 Feb 21;24(1):82. doi: 10.1186/s12864-023-09176-w. BMC Genomics. 2023. PMID: 36809964 Free PMC article. - Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress.
Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. Mittler R, et al. FEBS Lett. 2006 Dec 11;580(28-29):6537-42. doi: 10.1016/j.febslet.2006.11.002. Epub 2006 Nov 9. FEBS Lett. 2006. PMID: 17112521 Free PMC article. - Systemic and intracellular responses to photooxidative stress in Arabidopsis.
Rossel JB, Wilson PB, Hussain D, Woo NS, Gordon MJ, Mewett OP, Howell KA, Whelan J, Kazan K, Pogson BJ. Rossel JB, et al. Plant Cell. 2007 Dec;19(12):4091-110. doi: 10.1105/tpc.106.045898. Epub 2007 Dec 21. Plant Cell. 2007. PMID: 18156220 Free PMC article. - Comparative transcriptome profiling reveals cold stress responsiveness in two contrasting Chinese jujube cultivars.
Zhou H, He Y, Zhu Y, Li M, Song S, Bo W, Li Y, Pang X. Zhou H, et al. BMC Plant Biol. 2020 May 27;20(1):240. doi: 10.1186/s12870-020-02450-z. BMC Plant Biol. 2020. PMID: 32460709 Free PMC article.
References
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784 - PubMed
- Apel K, Hirt H (2004) reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 - PubMed
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B Methodological 57: 289–300
- Bent AF, Clough SJ (2000) Plant transformation. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–14
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases