Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes - PubMed (original) (raw)
Comparative Study
. 2005 Oct 11;102(41):14689-93.
doi: 10.1073/pnas.0506465102. Epub 2005 Oct 3.
Affiliations
- PMID: 16203998
- PMCID: PMC1239944
- DOI: 10.1073/pnas.0506465102
Comparative Study
Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes
Karsten Suhre et al. Proc Natl Acad Sci U S A. 2005.
Abstract
The initial analysis of the recently sequenced genome of Acanthamoeba polyphaga Mimivirus, the largest known double-stranded DNA virus, predicted a proteome of size and complexity more akin to small parasitic bacteria than to other nucleocytoplasmic large DNA viruses and identified numerous functions never before described in a virus. It has been proposed that the Mimivirus lineage could have emerged before the individualization of cellular organisms from the three domains of life. An exhaustive in silico analysis of the noncoding moiety of all known viral genomes now uncovers the unprecedented perfect conservation of an AAAATTGA motif in close to 50% of the Mimivirus genes. This motif preferentially occurs in genes transcribed from the predicted leading strand and is associated with functions required early in the viral infectious cycle, such as transcription and protein translation. A comparison with the known promoter of unicellular eukaryotes, amoebal protists in particular, strongly suggests that the AAAATTGA motif is the structural equivalent of the TATA box core promoter element. This element is specific to the Mimivirus lineage and may correspond to an ancestral promoter structure predating the radiation of the eukaryotic kingdoms. This unprecedented conservation of core promoter regions is another exceptional feature of Mimivirus that again raises the question of its evolutionary origin.
Figures
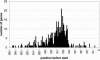
Fig. 1.
The distribution of the position of the AAAATTGA motif with respect to the predicted gene start shows the location of the conserved element at ≈50–110 nt.
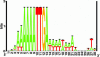
Fig. 2.
The sequence logo shows the conserved AT-rich neighborhood of the exactly conserved AAAATTGA octamer. The logo was based on 400 genes with a strictly conserved AAAATTGA motif; 3 genes with a motif that is <20 nt from the predicted translation start were not included in the computation of the logo. (See Fig. 5 for the corresponding logo of the
meme
motif.)
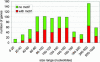
Fig. 3.
No relationship between the presence/absence of the AAAATTGA motif and the distance between two genes can be identified in the size distribution of Mimivirus intergenic regions. The exception is the virtual absence of the motif in intergenic regions that are too short to host it.
Fig. 4.
The alignment of the intergenic region of the paralogous gene cluster L175–L185 shows the perfect conservation of the AAAATTGA motif within intergenic regions that have otherwise more extensively diverged. Note that the AAAATTGA motif is part of the C terminus of gene L185 (indicated by white Xs).
Similar articles
- The 1.2-megabase genome sequence of Mimivirus.
Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. Raoult D, et al. Science. 2004 Nov 19;306(5700):1344-50. doi: 10.1126/science.1101485. Epub 2004 Oct 14. Science. 2004. PMID: 15486256 - Structural and functional insights into Mimivirus ORFans.
Saini HK, Fischer D. Saini HK, et al. BMC Genomics. 2007 May 9;8:115. doi: 10.1186/1471-2164-8-115. BMC Genomics. 2007. PMID: 17490476 Free PMC article. - Genomic and evolutionary aspects of Mimivirus.
Suzan-Monti M, La Scola B, Raoult D. Suzan-Monti M, et al. Virus Res. 2006 Apr;117(1):145-55. doi: 10.1016/j.virusres.2005.07.011. Epub 2005 Sep 21. Virus Res. 2006. PMID: 16181700 Review. - Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.
Iyer LM, Balaji S, Koonin EV, Aravind L. Iyer LM, et al. Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review. - Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae.
Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. Arslan D, et al. Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17486-91. doi: 10.1073/pnas.1110889108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987820 Free PMC article.
Cited by
- Preliminary crystallographic analysis of a polyadenylate synthase from Megavirus.
Lartigue A, Jeudy S, Bertaux L, Abergel C. Lartigue A, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Jan 1;69(Pt 1):53-6. doi: 10.1107/S1744309112048257. Epub 2012 Dec 20. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23295487 Free PMC article. - Locus-specific gene expression pattern suggests a unique propagation strategy for a giant algal virus.
Allen MJ, Forster T, Schroeder DC, Hall M, Roy D, Ghazal P, Wilson WH. Allen MJ, et al. J Virol. 2006 Aug;80(15):7699-705. doi: 10.1128/JVI.00491-06. J Virol. 2006. PMID: 16840348 Free PMC article. - Horizontal gene transfer and nucleotide compositional anomaly in large DNA viruses.
Monier A, Claverie JM, Ogata H. Monier A, et al. BMC Genomics. 2007 Dec 10;8:456. doi: 10.1186/1471-2164-8-456. BMC Genomics. 2007. PMID: 18070355 Free PMC article. - Mimiviridae: An Expanding Family of Highly Diverse Large dsDNA Viruses Infecting a Wide Phylogenetic Range of Aquatic Eukaryotes.
Claverie JM, Abergel C. Claverie JM, et al. Viruses. 2018 Sep 18;10(9):506. doi: 10.3390/v10090506. Viruses. 2018. PMID: 30231528 Free PMC article. Review. - Amoebae, Giant Viruses, and Virophages Make Up a Complex, Multilayered Threesome.
Diesend J, Kruse J, Hagedorn M, Hammann C. Diesend J, et al. Front Cell Infect Microbiol. 2018 Jan 11;7:527. doi: 10.3389/fcimb.2017.00527. eCollection 2017. Front Cell Infect Microbiol. 2018. PMID: 29376032 Free PMC article. Review.
References
- La Scola, B., Audic, S., Robert, C., Jungang, L., de Lamballerie, X., Drancourt, M., Birtles, R., Claverie, J.-M. & Raoult, D. (2003) Science 299, 2033. - PubMed
- Raoult, D., Audic, S., Robert, C., Abergel, C., Renesto, P., Ogata, H., La Scola, B., Suzan, M. & Claverie, J.-M. (2004) Science 306, 1344-1350. - PubMed
- Ogata, H., Audic, S., Renesto-Audiffren, P., Fournier, P. E., Barbe, V., Samson, D., Roux, V., Cossart, P., Weissenbach, J., Claverie, J.-M. & Raoult, D. (2001) Science 293, 2093-2098. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources