Defining the normal bacterial flora of the oral cavity - PubMed (original) (raw)
Comparative Study
Defining the normal bacterial flora of the oral cavity
Jørn A Aas et al. J Clin Microbiol. 2005 Nov.
Abstract
More than 700 bacterial species or phylotypes, of which over 50% have not been cultivated, have been detected in the oral cavity. Our purposes were (i) to utilize culture-independent molecular techniques to extend our knowledge on the breadth of bacterial diversity in the healthy human oral cavity, including not-yet-cultivated bacteria species, and (ii) to determine the site and subject specificity of bacterial colonization. Nine sites from five clinically healthy subjects were analyzed. Sites included tongue dorsum, lateral sides of tongue, buccal epithelium, hard palate, soft palate, supragingival plaque of tooth surfaces, subgingival plaque, maxillary anterior vestibule, and tonsils. 16S rRNA genes from sample DNA were amplified, cloned, and transformed into Escherichia coli. Sequences of 16S rRNA genes were used to determine species identity or closest relatives. In 2,589 clones, 141 predominant species were detected, of which over 60% have not been cultivated. Thirteen new phylotypes were identified. Species common to all sites belonged to the genera Gemella, Granulicatella, Streptococcus, and Veillonella. While some species were subject specific and detected in most sites, other species were site specific. Most sites possessed 20 to 30 different predominant species, and the number of predominant species from all nine sites per individual ranged from 34 to 72. Species typically associated with periodontitis and caries were not detected. There is a distinctive predominant bacterial flora of the healthy oral cavity that is highly diverse and site and subject specific. It is important to fully define the human microflora of the healthy oral cavity before we can understand the role of bacteria in oral disease.
Figures
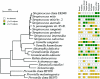
FIG. 1.
Bacterial profiles of the buccal epithelium of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are shown by the columns of boxes to the right of the tree as either not detected (clear box), <15% of the total number of clones assayed (shaded box), or ≥15% of the total number of clones assayed (darkened box). The 15% was chosen arbitrarily. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 2.
Bacterial profiles of the maxillary anterior vestibule of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 3.
Bacterial profiles of the tongue dorsum of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 4.
Bacterial profiles of the lateral tongue surface of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 5.
Bacterial profiles of the hard palate of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 6.
Bacterial profiles of the soft palate of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 7.
Bacterial profiles of the tonsils of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 8.
Bacterial profiles of the tooth surfaces of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 9.
Bacterial profiles of the subgingival plaque of healthy subjects. Distribution and levels of bacterial species/phylotypes among five subjects are as described for Fig. 1. Novel phylotypes identified in this study are indicated in bold. GenBank accession numbers are provided. Marker bar represents a 5% difference in nucleotide sequences.
FIG. 10.
Site specificity of predominant bacterial species in the oral cavity. In general, bacterial species or phylotypes were selected on the basis of their detection in multiple subjects for a given site. Distributions of bacterial species in oral sites among subjects are indicated by the columns of boxes to the right of the tree as follows: not detected in any subject (clear box), <15% of the total number of clones assayed (yellow box), ≥15% of the total number of clones assayed (green box). The 15% cutoff for low and high abundance was chosen arbitrarily. Marker bar represents a 10% difference in nucleotide sequences.
Similar articles
- Diversity and site-specificity of the oral microflora in the elderly.
Preza D, Olsen I, Willumsen T, Grinde B, Paster BJ. Preza D, et al. Eur J Clin Microbiol Infect Dis. 2009 Sep;28(9):1033-40. doi: 10.1007/s10096-009-0743-3. Epub 2009 Apr 17. Eur J Clin Microbiol Infect Dis. 2009. PMID: 19373498 Free PMC article. - Bacterial diversity in human subgingival plaque.
Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Paster BJ, et al. J Bacteriol. 2001 Jun;183(12):3770-83. doi: 10.1128/JB.183.12.3770-3783.2001. J Bacteriol. 2001. PMID: 11371542 Free PMC article. - Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients.
Kazor CE, Mitchell PM, Lee AM, Stokes LN, Loesche WJ, Dewhirst FE, Paster BJ. Kazor CE, et al. J Clin Microbiol. 2003 Feb;41(2):558-63. doi: 10.1128/JCM.41.2.558-563.2003. J Clin Microbiol. 2003. PMID: 12574246 Free PMC article. - Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories.
Woo PC, Lau SK, Teng JL, Tse H, Yuen KY. Woo PC, et al. Clin Microbiol Infect. 2008 Oct;14(10):908-34. doi: 10.1111/j.1469-0691.2008.02070.x. Clin Microbiol Infect. 2008. PMID: 18828852 Review. - Phylum Synergistetes in the oral cavity: A possible contributor to periodontal disease.
McCracken BA, Nathalia Garcia M. McCracken BA, et al. Anaerobe. 2021 Apr;68:102250. doi: 10.1016/j.anaerobe.2020.102250. Epub 2020 Aug 11. Anaerobe. 2021. PMID: 32791127 Review.
Cited by
- Gluten-degrading bacteria: availability and applications.
Kõiv V, Tenson T. Kõiv V, et al. Appl Microbiol Biotechnol. 2021 Apr;105(8):3045-3059. doi: 10.1007/s00253-021-11263-5. Epub 2021 Apr 10. Appl Microbiol Biotechnol. 2021. PMID: 33837830 Free PMC article. Review. - Microbial transformation from normal oral microbiota to acute endodontic infections.
Hsiao WW, Li KL, Liu Z, Jones C, Fraser-Liggett CM, Fouad AF. Hsiao WW, et al. BMC Genomics. 2012 Jul 28;13:345. doi: 10.1186/1471-2164-13-345. BMC Genomics. 2012. PMID: 22839737 Free PMC article. - Colonization, safety, and tolerability study of the Streptococcus salivarius 24SMBc nasal spray for its application in upper respiratory tract infections.
Santagati M, Scillato M, Muscaridola N, Metoldo V, La Mantia I, Stefani S. Santagati M, et al. Eur J Clin Microbiol Infect Dis. 2015 Oct;34(10):2075-80. doi: 10.1007/s10096-015-2454-2. Epub 2015 Jul 24. Eur J Clin Microbiol Infect Dis. 2015. PMID: 26205666 - Synergistic effects of arginine and fluoride on human dental biofilm control.
Kuriki N, Asahi Y, Okamoto M, Noiri Y, Ebisu S, Machi H, Suzuki M, Hayashi M. Kuriki N, et al. J Dent. 2024 Oct;149:105307. doi: 10.1016/j.jdent.2024.105307. Epub 2024 Aug 22. J Dent. 2024. PMID: 39178800 Free PMC article. - Oral microbiota diversity in moderate to severe plaque psoriasis, nail psoriasis and psoriatic arthritis.
Fan W, Lei N, Zheng Y, Liu J, Cao X, Su T, Su Z, Lu Y. Fan W, et al. Sci Rep. 2024 Aug 8;14(1):18402. doi: 10.1038/s41598-024-69132-w. Sci Rep. 2024. PMID: 39117753 Free PMC article.
References
- Albandar, J. M., J. A. Brunelle, and A. Kingman. 1999. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988-1994. J. Periodontol. 70:13-29. - PubMed
- Beck, J., R. Garcia, G. Heiss, P. S. Vokonas, and S. Offenbacher. 1996. Periodontal disease and cardiovascular disease. J. Periodontol. 67:1123-1137. - PubMed
- Berbari, E. F., F. R. Cockerill III, and J. M. Steckelberg. 1997. Infective endocarditis due to unusual or fastidious microorganisms. Mayo Clin. Proc. 72:532-542. - PubMed
- Bouvet, A., and J. F. Acar. 1984. New bacteriological aspects of infective endocarditis. Eur. Heart J. 5(Suppl. C):45-48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases