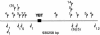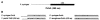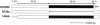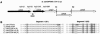Yersiniabactin production by Pseudomonas syringae and Escherichia coli, and description of a second yersiniabactin locus evolutionary group - PubMed (original) (raw)
Yersiniabactin production by Pseudomonas syringae and Escherichia coli, and description of a second yersiniabactin locus evolutionary group
Alain Bultreys et al. Appl Environ Microbiol. 2006 Jun.
Abstract
The siderophore and virulence factor yersiniabactin is produced by Pseudomonas syringae. Yersiniabactin was originally detected by high-pressure liquid chromatography (HPLC); commonly used PCR tests proved ineffective. Yersiniabactin production in P. syringae correlated with the possession of irp1 located in a predicted yersiniabactin locus. Three similarly divergent yersiniabactin locus groups were determined: the Yersinia pestis group, the P. syringae group, and the Photorhabdus luminescens group; yersiniabactin locus organization is similar in P. syringae and P. luminescens. In P. syringae pv. tomato DC3000, the locus has a high GC content (63.4% compared with 58.4% for the chromosome and 60.1% and 60.7% for adjacent regions) but it lacks high-pathogenicity-island features, such as the insertion in a tRNA locus, the integrase, and insertion sequence elements. In P. syringae pv. tomato DC3000 and pv. phaseolicola 1448A, the locus lies between homologues of Psyr_2284 and Psyr_2285 of P. syringae pv. syringae B728a, which lacks the locus. Among tested pseudomonads, a PCR test specific to two yersiniabactin locus groups detected a locus in genospecies 3, 7, and 8 of P. syringae, and DNA hybridization within P. syringae also detected a locus in the pathovars phaseolicola and glycinea. The PCR and HPLC methods enabled analysis of nonpathogenic Escherichia coli. HPLC-proven yersiniabactin-producing E. coli lacked modifications found in irp1 and irp2 in the human pathogen CFT073, and it is not clear whether CFT073 produces yersiniabactin. The study provides clues about the evolution and dispersion of yersiniabactin genes. It describes methods to detect and study yersiniabactin producers, even where genes have evolved.
Figures
FIG. 1.
Detection (A) in GASN medium with the HPLC program 2 of Fe-PVD and Fe-YBT produced by P. syringae pv. tomato LMG 5093 and (B) in King's medium B with the HPLC program 3 of Fe-YBT produced by E. coli ECOR 10. For each strain, an HPLC analysis (on the left) and the spectral characteristics of Fe-YBT analyzed in line (on the right) are shown. Both HPLC programs can be used for each species. In King's medium B (B), Fe-YBT is more easily detected at 403 nm because medium components (visible between 2 and 10 min) absorb more at 305 nm. (C) ESI-MS positive-ion analysis of Fe-YBT of P. syringae pv. tomato LMG 5093.
FIG. 2.
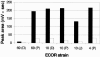
YBT production in King's medium B by the E. coli strains ECOR 69, ECOR 10, and ECOR 4 received from different origins: (P), received from B. Picard; (D), received from E. Denamur; and (J), received from J. R. Johnson (Table 2). The strains were grown at 37°C for 2 days in either one still petri dish with one agar block (black bars) or one shaken Erlenmeyer flask (white bars). YBT production was assessed by the Fe-YBT HPLC peak area at 403 nm.
FIG. 3.
Map of the YBT locus in (A) Y. pestis 91001, (B) P. syringae DC3000, and (C) P. luminescens TTO1, according to annotated genomes. The genes are designed according to the encoded protein functions: black, biosynthetic enzymes, apart from ybtS homologues, represented by a dark squaring; gray, membrane receptors; horizontal lines, transport proteins; light squaring, regulatory proteins; and vertical lines, proteins of unknown function. The white-coded genes have no homologues in the other species.
FIG. 4.
Map and orientation of IS elements around the YBT locus (black box) of P. syringae DC3000 according to the annotated genome: 1, IS_Pssy_; 4, IS_Psy4_; 5, IS_Psy5_; 6, IS_Psy6_; 7, IS_Psy7_; 12, IS_Psy12_; 14, IS_Psy14_. The parentheses indicate disrupted IS elements.
FIG. 5.
(A) Location of the chorismate binding domain (CBD, black bar) detected in PchA of P. syringae DC3000 (gray bar) using the NCBI conserved domain search. (B) BLAST2 Sequences comparison of PchA of P. syringae DC3000 with either YbtS of Y. pestis 91001 or PchA of P. aeruginosa PAO1; the proteins are represented by thin lines, apart from zones with high similarity, which are represented by wide gray bars, and gaps, represented by intermediate-thickness dark gray bars.
FIG. 6.
Homologous regions around the YBT locus in P. syringae pv. tomato DC3000, P. syringae pv. syringae B728a, and P. syringae pv. phaseolicola 1448A and GC contents (P. syringae pv. tomato DC3000). Homologous regions are represented by rectangles; the YBT loci and the adjacent gene PSPTO2607 (DC3000) or PSPPH2892 (1448A) are black coded. Nonhomologous regions are shown either by a dark line when homologous proteins were found elsewhere in the genome of another strain or by a gray line when homologous proteins were not found anywhere in the genome of either of the other strains. The dotted line represents a gap. Homologous regions were determined by comparing protein or gene sequences using SSearch, BLASTP, or BLASTN and by comparing gene positions; protein homologies observed were higher than 80% identity. The last left and right genes represented that have a homologue in the same area in another pathovar are PSPTO2573 and PSPTO2609 (DC3000), Psyr_2264 and Psyr_2289 (B728a), and PSPPH2922 and PSPPH2887 (1448A); in 1448A, PSPPH2923 is an IS_801_ transposase.
FIG. 7.
(A) Map of the irp2 and irp1 homologous zones in E. coli CFT073, as annotated in its genome. Shown in the irp2 zone are two ORFs (black arrows), a 711-bp IS_1541A_-like due insertion (IS), and the primers used in PCR analyses. Shown in the irp1 zone are three ORFs (white arrows) and the modified segments S1 and S2. (B) Comparisons of S1 and S2 in Y. pestis and E. coli. In E. coli CFT073, modifications (gray) induce the lecture of stop codons (bold); in the next ORFs, the correct translation would restart downstream of the last base modification. No indication of this was found in the six YBT-producing ECOR strains, which indicates that irp1 encodes HMWP1 in these strains.
Similar articles
- Characterization of pyoverdine and achromobactin in Pseudomonas syringae pv. phaseolicola 1448a.
Owen JG, Ackerley DF. Owen JG, et al. BMC Microbiol. 2011 Oct 3;11:218. doi: 10.1186/1471-2180-11-218. BMC Microbiol. 2011. PMID: 21967163 Free PMC article. - Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000.
Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A, Lykidis A, Trong S, Nolan M, Goltsman E, Thiel J, Malfatti S, Loper JE, Lapidus A, Detter JC, Land M, Richardson PM, Kyrpides NC, Ivanova N, Lindow SE. Feil H, et al. Proc Natl Acad Sci U S A. 2005 Aug 2;102(31):11064-9. doi: 10.1073/pnas.0504930102. Epub 2005 Jul 25. Proc Natl Acad Sci U S A. 2005. PMID: 16043691 Free PMC article. - Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains.
Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A. Lindeberg M, et al. Mol Plant Microbe Interact. 2006 Nov;19(11):1151-8. doi: 10.1094/MPMI-19-1151. Mol Plant Microbe Interact. 2006. PMID: 17073298 Review. - Roadmap to new virulence determinants in Pseudomonas syringae: insights from comparative genomics and genome organization.
Lindeberg M, Myers CR, Collmer A, Schneider DJ. Lindeberg M, et al. Mol Plant Microbe Interact. 2008 Jun;21(6):685-700. doi: 10.1094/MPMI-21-6-0685. Mol Plant Microbe Interact. 2008. PMID: 18624633 Review.
Cited by
- Siderophores in environmental research: roles and applications.
Ahmed E, Holmström SJ. Ahmed E, et al. Microb Biotechnol. 2014 May;7(3):196-208. doi: 10.1111/1751-7915.12117. Epub 2014 Feb 27. Microb Biotechnol. 2014. PMID: 24576157 Free PMC article. Review. - Wide Distribution of Foxicin Biosynthetic Gene Clusters in Streptomyces Strains - An Unusual Secondary Metabolite with Various Properties.
Greule A, Marolt M, Deubel D, Peintner I, Zhang S, Jessen-Trefzer C, De Ford C, Burschel S, Li SM, Friedrich T, Merfort I, Lüdeke S, Bisel P, Müller M, Paululat T, Bechthold A. Greule A, et al. Front Microbiol. 2017 Feb 21;8:221. doi: 10.3389/fmicb.2017.00221. eCollection 2017. Front Microbiol. 2017. PMID: 28270798 Free PMC article. - Data-Independent Acquisition Proteomics Unravels the Effects of Iron Ions on Coronatine Synthesis in Pseudomonas syringae pv. tomato DC3000.
He Y, Yu S, Liu S, Tian H, Yu C, Tan W, Zhang J, Li Z, Jiang F, Duan L. He Y, et al. Front Microbiol. 2020 Jul 21;11:1362. doi: 10.3389/fmicb.2020.01362. eCollection 2020. Front Microbiol. 2020. PMID: 32793123 Free PMC article. - Iron-regulated metabolites produced by Pseudomonas fluorescens WCS374r are not required for eliciting induced systemic resistance against Pseudomonas syringae pv. tomato in Arabidopsis.
Djavaheri M, Mercado-Blanco J, Versluis C, Meyer JM, Loon LC, Bakker PA. Djavaheri M, et al. Microbiologyopen. 2012 Sep;1(3):311-25. doi: 10.1002/mbo3.32. Epub 2012 Aug 24. Microbiologyopen. 2012. PMID: 23170230 Free PMC article.
References
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidlan, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- Bach, S., A. de Almeida, and E. Carniel. 2000. The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183:289-294. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous