Passive sexual transmission of human immunodeficiency virus type 1 variants and adaptation in new hosts - PubMed (original) (raw)
Comparative Study
. 2006 Jul;80(14):7226-34.
doi: 10.1128/JVI.02014-05.
C T T Edwards, N McCarthy, J Fox, H Brown, A Milicic, N Mackie, T Pillay, J W Drijfhout, S Dustan, J R Clarke, E C Holmes, H T Zhang, K Pfafferott, P J Goulder, M O McClure, J Weber, R E Phillips, S Fidler
Affiliations
- PMID: 16809328
- PMCID: PMC1489048
- DOI: 10.1128/JVI.02014-05
Comparative Study
Passive sexual transmission of human immunodeficiency virus type 1 variants and adaptation in new hosts
A J Frater et al. J Virol. 2006 Jul.
Abstract
Human immunodeficiency virus type 1 (HIV-1) genetic diversity is a major obstacle for the design of a successful vaccine. Certain viral polymorphisms encode human leukocyte antigen (HLA)-associated immune escape, potentially overcoming limited vaccine protection. Although transmission of immune escape variants has been reported, the overall extent to which this phenomenon occurs in populations and the degree to which it contributes to HIV-1 viral evolution are unknown. Selection on the HIV-1 env gene at transmission favors neutralization-sensitive variants, but it is not known to what degree selection acts on the internal HIV-1 proteins to restrict or enhance the transmission of immune escape variants. Studies have suggested that HLA class I may determine susceptibility to HIV-1 infection, but a definitive role for HLA at transmission remains unproven. Comparing populations of acute seroconverters and chronically infected patients, we found no evidence of selection acting to restrict transmission of HIV-1 variants. We found that statistical associations previously reported in chronic infection between viral polymorphisms and HLA class I alleles are not present in acute infection, suggesting that the majority of viral polymorphisms in these patients are the result of transmission rather than de novo adaptation. Using four episodes of HIV-1 transmission in which the donors and recipients were both sampled very close to the time of infection we found that, despite a transmission bottleneck, genetic variants of HIV-1 infection are transmitted in a frequency-dependent manner. As HIV-1 infections are seeded by unique donor-adapted viral variants, each episode is a highly individual antigenic challenge. Host-specific, idiosyncratic HIV-1 antigenic diversity will seriously tax the efficacy of immunization based on consensus sequences.
Figures
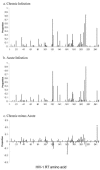
FIG. 1.
Population-level distribution of amino acid variation in RT in two patient cohorts. Variant frequency in the HIV-1 RT gene (amino acids 1 to 250) across a population of 62 drug-naïve chronically infected patients with subtype B HIV-1 recruited at St. Mary's Hospital, London (a) and a population of 101 individuals sampled shortly after infection recruited at St. Mary's Hospital, London (b). (c) Differences in polymorphism frequencies for the HIV-1 RT gene in acute and chronic patients. *, site at which the difference in proportions is statistically significant.
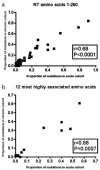
FIG. 2.
Analyses of variant frequencies in acute versus chronic patients using Spearman's nonparametric correlation coefficient for all polymorphic sites in RT (a) or only sites at which variation is associated with the presence of the restricting HLA class I allele (b). Correlations are shown between acute seroconverters from St. Mary's Hospital and chronically infected patients from St. Mary's Hospital.
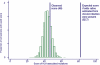
FIG. 3.
HLA class I-independent variation in acutely infected patients. For each site known to adapt to the class I-restricted CTL response (see Table S1 in the supplemental material), the number of times a variant was observed in association with an HLA allele known to direct variation at that site was counted. The observed score for the seroconverter cohort was 46. Patient sequences were then randomly assigned to different HLA class I alleles expressed in the seroconverter cohort. Repetition of this process generated the null distribution of scores expected if the amino acid variants expressed by an infecting virus were unrelated to the HLA type of the patient. The mean for the null distribution was 42.4. The expected score for a cohort of previously described chronically infected patients (calculated using published odds ratios) (21) is 82.7, which is significantly different from the null distribution (P < 0.0001).
FIG. 4.
(a) Neighbor-joining phylogenetic tree of gag p24 nucleotide sequences from donor-recipient pairs, assuming the HKY85 model of nucleotide substitution. Sequences were 843 bp long. The number of sequences for each patient was as follows: D1, 48; R1, 43; D2, 19; R2, 14; D3, 29; R3, 25; D4, 38; R4, 27. Subtype A and B reference sequences were obtained from the HIV Sequence Database (
). Horizontal branch lengths are drawn to scale. Divergence is the number of substitutions per site, expressed as a percentage. (b) Retention of HIV-1 amino acid variants during sexual transmission. gag p24 from four donor-recipient pairs is shown. The variant frequency (proportion of amino acids at a particular site that differ from the subtype consensus sequence) is plotted on the vertical axis. Different-colored bars represent different amino acids at a given site. The horizontal axis represents the linear amino acid sequence from position 1 to 278. Regions of gag p24 restricted by the HLA alleles expressed in the infected patient are marked by red bars.
FIG. 5.
Frequency-dependent transmission of HLA class I antigenic variants, as shown by donor frequencies of transmission of all intraepitope, polymorphic sites. The median frequencies for the transmitted and nontransmitted variants are shown and were found to be significantly different (P < 0.0001, two-tailed Mann-Whitney test).
Similar articles
- Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1.
Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, Kadie C, Bhattacharya T, Chui C, Szinger J, Mo T, Hogg RS, Montaner JS, Frahm N, Brander C, Walker BD, Harrigan PR. Brumme ZL, et al. PLoS Pathog. 2007 Jul;3(7):e94. doi: 10.1371/journal.ppat.0030094. PLoS Pathog. 2007. PMID: 17616974 Free PMC article. - Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA.
Leslie A, Kavanagh D, Honeyborne I, Pfafferott K, Edwards C, Pillay T, Hilton L, Thobakgale C, Ramduth D, Draenert R, Le Gall S, Luzzi G, Edwards A, Brander C, Sewell AK, Moore S, Mullins J, Moore C, Mallal S, Bhardwaj N, Yusim K, Phillips R, Klenerman P, Korber B, Kiepiela P, Walker B, Goulder P. Leslie A, et al. J Exp Med. 2005 Mar 21;201(6):891-902. doi: 10.1084/jem.20041455. J Exp Med. 2005. PMID: 15781581 Free PMC article. - Rare HLA drive additional HIV evolution compared to more frequent alleles.
Rousseau CM, Lockhart DW, Listgarten J, Maley SN, Kadie C, Learn GH, Nickle DC, Heckerman DE, Deng W, Brander C, Ndung'u T, Coovadia H, Goulder PJ, Korber BT, Walker BD, Mullins JI. Rousseau CM, et al. AIDS Res Hum Retroviruses. 2009 Mar;25(3):297-303. doi: 10.1089/aid.2008.0208. AIDS Res Hum Retroviruses. 2009. PMID: 19327049 Free PMC article. - Clinical and evolutionary consequences of HIV adaptation to HLA: implications for vaccine and cure.
Avila-Rios S, Carlson JM, John M, Mallal S, Brumme ZL. Avila-Rios S, et al. Curr Opin HIV AIDS. 2019 May;14(3):194-204. doi: 10.1097/COH.0000000000000541. Curr Opin HIV AIDS. 2019. PMID: 30925534 Free PMC article. Review. - Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence.
Burns DP, Desrosiers RC. Burns DP, et al. Curr Top Microbiol Immunol. 1994;188:185-219. doi: 10.1007/978-3-642-78536-8_11. Curr Top Microbiol Immunol. 1994. PMID: 7523031 Review.
Cited by
- Epigenetic Features of HIV-Induced T-Cell Exhaustion Persist Despite Early Antiretroviral Therapy.
Martin GE, Sen DR, Pace M, Robinson N, Meyerowitz J, Adland E, Thornhill JP, Jones M, Ogbe A, Parolini L, Olejniczak N, Zacharopoulou P, Brown H, Willberg CB, Nwokolo N, Fox J, Fidler S, Haining WN, Frater J. Martin GE, et al. Front Immunol. 2021 Jun 4;12:647688. doi: 10.3389/fimmu.2021.647688. eCollection 2021. Front Immunol. 2021. PMID: 34149690 Free PMC article. Clinical Trial. - [ADS-J1 antagonizes semen-derived enhancer of virus infection-mediated enhancement of transmitted founder HIV-1 and its matched chronic control strain infection].
Liu HM, Ma NN, Luo C, Yuan SY, Liu FL, Yao XG, Zhou CQ, Zou M. Liu HM, et al. Nan Fang Yi Ke Da Xue Xue Bao. 2018 Feb 20;38(2):211-216. doi: 10.3969/j.issn.1673-4254.2018.02.15. Nan Fang Yi Ke Da Xue Xue Bao. 2018. PMID: 29502062 Free PMC article. Chinese. - Post-treatment control or treated controllers? Viral remission in treated and untreated primary HIV infection.
Martin GE, Gossez M, Williams JP, Stöhr W, Meyerowitz J, Leitman EM, Goulder P, Porter K, Fidler S, Frater J; the SPARTAC Trial Investigators. Martin GE, et al. AIDS. 2017 Feb 20;31(4):477-484. doi: 10.1097/QAD.0000000000001382. AIDS. 2017. PMID: 28060012 Free PMC article. Clinical Trial. - Exceptional Heterogeneity in Viral Evolutionary Dynamics Characterises Chronic Hepatitis C Virus Infection.
Raghwani J, Rose R, Sheridan I, Lemey P, Suchard MA, Santantonio T, Farci P, Klenerman P, Pybus OG. Raghwani J, et al. PLoS Pathog. 2016 Sep 15;12(9):e1005894. doi: 10.1371/journal.ppat.1005894. eCollection 2016 Sep. PLoS Pathog. 2016. PMID: 27631086 Free PMC article. - HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck.
Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, Lin TH, Peng J, Seese AM, Shapiro R, Frater J, Ndung'u T, Tang J, Goepfert P, Gilmour J, Price MA, Kilembe W, Heckerman D, Goulder PJ, Allen TM, Allen S, Hunter E. Carlson JM, et al. Science. 2014 Jul 11;345(6193):1254031. doi: 10.1126/science.1254031. Epub 2014 Jul 10. Science. 2014. PMID: 25013080 Free PMC article.
References
- Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. - PMC - PubMed
- Allen, T. M., M. Altfeld, X. G. Yu, K. M. O'Sullivan, M. Lichterfeld, S. Le Gall, M. John, B. R. Mothe, P. K. Lee, E. T. Kalife, D. E. Cohen, K. A. Freedberg, D. A. Strick, M. N. Johnston, A. Sette, E. S. Rosenberg, S. A. Mallal, P. J. Goulder, C. Brander, and B. D. Walker. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 78:7069-7078. - PMC - PubMed
- Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. - PubMed
- Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G108/626/MRC_/Medical Research Council/United Kingdom
- R01 AI046995/AI/NIAID NIH HHS/United States
- AI046995-06/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials