Transmembrane segment 7 of human P-glycoprotein forms part of the drug-binding pocket - PubMed (original) (raw)
Transmembrane segment 7 of human P-glycoprotein forms part of the drug-binding pocket
Tip W Loo et al. Biochem J. 2006.
Abstract
P-gp (P-glycoprotein; ABCB1) protects us by transporting a broad range of structurally unrelated compounds out of the cell. Identifying the regions of P-gp that make up the drug-binding pocket is important for understanding the mechanism of transport. The common drug-binding pocket is at the interface between the transmembrane domains of the two homologous halves of P-gp. It has been shown in a previous study [Loo, Bartlett and Clarke (2006) Biochem. J. 396, 537-545] that the first transmembrane segment (TM1) contributed to the drug-binding pocket. In the present study, we used cysteine-scanning mutagenesis, reaction with an MTS (methanethiosulfonate) thiol-reactive analogue of verapamil (termed MTS-verapamil) and cross-linking analysis to test whether the equivalent transmembrane segment (TM7) in the C-terminal-half of P-gp also contributed to drug binding. Mutation of Phe728 to cysteine caused a 4-fold decrease in apparent affinity for the drug substrate verapamil. Mutant F728C also showed elevated ATPase activity (11.5-fold higher than untreated controls) after covalent modification with MTS-verapamil. The activity returned to basal levels after treatment with dithiothreitol. The substrates, verapamil and cyclosporin A, protected the mutant from labelling with MTS-verapamil. Mutant F728C could be cross-linked with a homobifunctional thiol-reactive cross-linker to cysteines I306C(TM5) and F343C(TM6) that are predicted to line the drug-binding pocket. Disulfide cross-linking was inhibited by some drug substrates such as Rhodamine B, calcein acetoxymethyl ester, cyclosporin, verapamil and vinblastine or by vanadate trapping of nucleotides. These results indicate that TM7 forms part of the drug-binding pocket of P-gp.
Figures
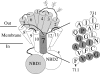
Figure 1. Schematic models of P-gp and composition of TM7
The 12 TMs of P-gp are shown as numbered cylinders. The branched lines represent glycosylation sites. NBD1 and NBD2 represent the nucleotide-binding domains. The amino acids in predicted TM7 (positions 711–731) are shown as an α-helical net. The amino acids identical with those in the predicted TM1 structure are shaded in grey.
Figure 2. Effect of MTS–verapamil on basal ATPase activity of TM7 cysteine mutants
(A) His-tagged Cys-less or mutants F711C/I731C P-gps were expressed in HEK-293 cells and solubilized with _n_-dodecyl-β-
D
-maltoside. Insoluble material was removed by centrifugation and the supernatants were treated with or without 0.3 mM MTS–verapamil. His-tagged P-gps were then isolated by nickel-chelate chromatography. Equivalent amounts of P-gp were mixed with lipid, sonicated and assayed for ATPase activity in the absence of drug substrate. ND, not determined because of low expression. (B) Cys-less, Q725C, F728C and A729C P-gp mutants were treated with or without 3 mM MTS–verapamil and His-tagged P-gp isolated by nickel-chelate chromatography. Equivalent amounts of P-gp were mixed with lipid and ATPase activity determined in the presence (+) or absence (−) of 1 mM verapamil (Ver). The fold-stimulation is the ratio of activity of a sample treated with MTS–verapamil to that of an untreated sample. Each value is the mean±S.D. (_n_=3).
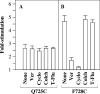
Figure 3. Inhibition of MTS–verapamil labelling of mutants Q725C and F728C by substrates
HEK-293 cells expressing mutants Q725C or F728C were solubilized with _n_-dodecyl-β-
D
-maltoside. Insoluble material was removed by centrifugation. Equivalent amounts of supernatant were incubated for 10 min at 20 °C in the presence of no drug (None), 3 mM verapamil (Ver), 0.2 mM cyclosporin A (Cyclo), 10 mM colchicine (Colch) or 0.6 mM _trans_-(E)-flupentixol (T-Flu). The mixtures were then incubated with or without 0.3 mM MTS–verapamil for 10 min at 20 °C. His-tagged P-gps were isolated by nickel-chelate chromatography, mixed with an equal volume of lipid and assayed for ATPase activity. The activities are expressed relative to that of a sample not treated with MTS–verapamil. Each value is the mean±S.D. (_n_=3).
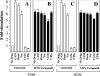
Figure 4. Effect of drug substrates and inhibitors on the ATPase activity of mutants F728C and Q725C before and after labelling with MTS–verapamil
HEK-293 cells expressing His-tagged mutants F728C or Q725C were solubilized with _n_-dodecyl-β-
D
-maltoside and then incubated in the absence (A and C) or presence (B and D) of 3 mM MTS–verapamil. P-gp was then isolated by nickel-chelate chromatography, mixed with E. coli lipids and ATPase activity measured in the absence (No Drug) or presence of 0.6 mM calcein–AM (CAM), 3 mM demecolcine (Deme), 1 mM verapamil (Ver), 0.2 mM cyclosporin A (Cyclo) or 0.6 mM _trans_-(E)-flupentixol (T-Flu). The fold-stimulation is the ratio of activity with drug substrate to that of the unlabelled sample assayed without drug substrate. Each value is the mean±S.D. (_n_=3).
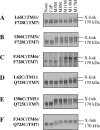
Figure 5. Disulfide cross-linking of P-gp mutants
Membranes prepared from HEK-293 cells expressing mutants (A) L65C(TM1)/F728C(TM7), (B) I306C(TM5)/F728C(TM7), (C) F343C(TM6)/F728C(TM7), (D) L65C(TM1)/Q725C(TM7), (E) I306C(TM5)/Q725C(TM7) or (F) F343C(TM6)/Q725C(TM7) were treated with 1 mM copper phenanthroline (CuP), 0.2 mM of the homobifunctional disulfide cross-linker M5M, M8M, M11M, M14M or M17M, or no cross-linker (None) for 15 min at 4 °C. The reactions were stopped by addition of SDS sample buffer containing no reducing agent, and samples were subjected to immunoblot analysis on SDS/PAGE (7.5% gels). The positions of the cross-linked (X-link) and mature (170 kDa) P-gps are indicated.
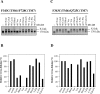
Figure 6. Effect of substrates on cross-linking of mutants F343C(TM6)/F728C(TM7) or F343C(TM6)/Q725C(TM7)
(A and C) Membranes prepared from HEK-293 cells expressing mutants F343C(TM6)/F728C(TM7) or F343C(TM6)/Q725C(TM7) were pre-incubated for 10 min at 20 °C with no drug (None), 10 mM colchicine (Colch), 0.05 mM cyclosporin A (Cyclo), 2 mM Rhodamine B (Rhod), 1 mM MTS–verapamil (Ver), 0.2 mM vinblastine (Vin), 3 mM demecolcine (Deme), 0.3 mM Hoechst 33342 (Hoech), 0.6 mM _trans_-(E)-flupentixol (T-Flu), 0.6 mM cis-(Z)-flupentixol (C-Flu) or 0.6 mM calcein–AM (Calcein). The samples were cooled to 4 °C and then treated with (+) or without (−) 0.2 mM M14M cross-linker for 2 min at 4 °C. The reactions were stopped by addition of SDS sample buffer containing no reducing agent, and samples were subjected to immunoblot analysis on SDS/PAGE (7.5% gels). The positions of the cross-linked (X-link) and mature (170 kDa) P-gps are indicated. (B and D) The relative amount of cross-linking was determined by scanning the gel lanes, and the results expressed as the amount of cross-linked product in the presence of drug substrate to that cross-linked with no drug substrate present (None taken as 100%).
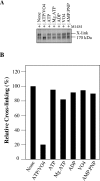
Figure 7. Effect of vanadate trapping of nucleotide on cross-linking of mutant F343C(TM6)/F728C(TM7)
(A) Membranes prepared from HEK-293 cells expressing mutant F343C(TM6)/F728C(TM7) were pre-incubated for 20 min at 4 °C with no addition (None) or with 5 mM ATP (ATP), 10 mM MgCl2 and 5 mM ATP (Mg.ATP), 10 mM MgCl2 and 5 mM ADP (ADP), 10 mM MgCl2 and 0.2 mM sodium orthovanadate (VO4) or 10 mM MgCl2 and 5 mM adenosine 5′-[β, γ-imino]triphosphate (AMP.PNP). One sample was incubated with 10 mM MgCl2, 5 mM ATP and 0.2 mM sodium orthovanadate (ATP/VO4) at 37 °C for 10 min to convert P-gp into the vanadate-trapped transition state. The samples were cooled to 4 °C and then treated with (+) 0.2 mM M14M cross-linker for 2 min at 4 °C. The reactions were stopped by addition of SDS sample buffer containing no reducing agent, and samples were subjected to immunoblot analysis on SDS/PAGE (7.5% gels). The positions of the cross-linked (X-link) and mature (170 kDa) P-gps are indicated. (B) The relative amount of cross-linking was determined by scanning the gel lanes, and the results expressed as the amount of cross-linked product in the presence of drug substrate to that cross-linked with no drug substrate present (None taken as 100%).
Figure 8. Residues in TM1 and TM7 arranged as α-helices
The residues of TM1 and TM7 are arranged as cylindrical helices and alignment is based on amino acid identity. Residues that show alterations in their ATPase activity after reaction with MTS–verapamil or show disulfide cross-linking are shown occupying one face of the helix (enclosed in a large circle). Black circles indicate the positions of residues that are fully activated after reaction with MTS–verapamil and whose labelling by MTS–verapamil is blocked by drug substrate. The dark grey circle indicates the position of a cysteine that is highly activated by MTS–verapamil, but whose labelling by MTS–verapamil is not protected by drug substrate. The light grey circles indicate the positions of cysteines that show inhibition of verapamil-stimulated (729) or basal ATPase activity (726) after reaction with MTS–verapamil. Residues 68 and 69 in TM1 (in bold) when changed to cysteine could be cross-linked to cysteines in TM11 during ATP hydrolysis [43].
Similar articles
- Transmembrane segment 1 of human P-glycoprotein contributes to the drug-binding pocket.
Loo TW, Bartlett MC, Clarke DM. Loo TW, et al. Biochem J. 2006 Jun 15;396(3):537-45. doi: 10.1042/BJ20060012. Biochem J. 2006. PMID: 16492138 Free PMC article. - The drug-binding pocket of the human multidrug resistance P-glycoprotein is accessible to the aqueous medium.
Loo TW, Bartlett MC, Clarke DM. Loo TW, et al. Biochemistry. 2004 Sep 28;43(38):12081-9. doi: 10.1021/bi049045t. Biochemistry. 2004. PMID: 15379547 - The power of the pump: mechanisms of action of P-glycoprotein (ABCB1).
Ambudkar SV, Kim IW, Sauna ZE. Ambudkar SV, et al. Eur J Pharm Sci. 2006 Apr;27(5):392-400. doi: 10.1016/j.ejps.2005.10.010. Epub 2005 Dec 13. Eur J Pharm Sci. 2006. PMID: 16352426 Review. - How does P-glycoprotein recognize its substrates?
Ueda K, Taguchi Y, Morishima M. Ueda K, et al. Semin Cancer Biol. 1997 Jun;8(3):151-9. doi: 10.1006/scbi.1997.0066. Semin Cancer Biol. 1997. PMID: 9441945 Review.
Cited by
- Characterizing the binding interactions between P-glycoprotein and eight known cardiovascular transport substrates.
Jagodinsky JC, Akgun U. Jagodinsky JC, et al. Pharmacol Res Perspect. 2015 Mar;3(2):e00114. doi: 10.1002/prp2.114. Pharmacol Res Perspect. 2015. PMID: 25729581 Free PMC article. - Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans.
Jin MS, Oldham ML, Zhang Q, Chen J. Jin MS, et al. Nature. 2012 Oct 25;490(7421):566-9. doi: 10.1038/nature11448. Epub 2012 Sep 23. Nature. 2012. PMID: 23000902 Free PMC article. - Targeted degradation of ABC transporters in health and disease.
Nikles D, Tampé R. Nikles D, et al. J Bioenerg Biomembr. 2007 Dec;39(5-6):489-97. doi: 10.1007/s10863-007-9120-z. J Bioenerg Biomembr. 2007. PMID: 17972020 Review. - Residues contributing to drug transport by ABCG2 are localised to multiple drug-binding pockets.
Cox MH, Kapoor P, Briggs DA, Kerr ID. Cox MH, et al. Biochem J. 2018 May 4;475(9):1553-1567. doi: 10.1042/BCJ20170923. Biochem J. 2018. PMID: 29661915 Free PMC article. - ABCB transporters in a leaf beetle respond to sequestered plant toxins.
Kowalski P, Baum M, Körten M, Donath A, Dobler S. Kowalski P, et al. Proc Biol Sci. 2020 Sep 9;287(1934):20201311. doi: 10.1098/rspb.2020.1311. Epub 2020 Sep 2. Proc Biol Sci. 2020. PMID: 32873204 Free PMC article.
References
- Holland I. B., Blight M. A. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 1999;293:381–399. - PubMed
- Ambudkar S. V., Dey S., Hrycyna C. A., Ramachandra M., Pastan I., Gottesman M. M. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu. Rev. Pharmacol. Toxicol. 1999;39:361–398. - PubMed
- Schinkel A. H., Smit J. J., van Tellingen O., Beijnen J. H., Wagenaar E., van Deemter L., Mol C. A., van der Valk M. A., Robanus-Maandag E. C., te Riele H. P., Berns A. J. M., Borst P. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77:491–502. - PubMed
- Lee C. G., Gottesman M. M., Cardarelli C. O., Ramachandra M., Jeang K. T., Ambudkar S. V., Pastan I., Dey S. HIV-1 protease inhibitors are substrates for the MDR1 multidrug transporter. Biochemistry. 1998;37:3594–3601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous