Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans - PubMed (original) (raw)
Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans
Aysha H Osmani et al. Mol Biol Cell. 2006 Dec.
Abstract
To define the extent of the modification of the nuclear pore complex (NPC) during Aspergillus nidulans closed mitosis, a systematic analysis of nuclear transport genes has been completed. Thirty genes have been deleted defining 12 nonessential and 18 essential genes. Several of the nonessential deletions caused conditional phenotypes and self-sterility, whereas deletion of some essential genes caused defects in nuclear structure. Live cell imaging of endogenously tagged NPC proteins (Nups) revealed that during mitosis 14 predicted peripheral Nups, including all FG repeat Nups, disperse throughout the cell. A core mitotic NPC structure consisting of membrane Nups, all components of the An-Nup84 subcomplex, An-Nup170, and surprisingly, An-Gle1 remained throughout mitosis. We propose this minimal mitotic NPC core provides a conduit across the nuclear envelope and acts as a scaffold to which dispersed Nups return during mitotic exit. Further, unlike other dispersed Nups, An-Nup2 locates exclusively to mitotic chromatin, suggesting it may have a novel mitotic role in addition to its nuclear transport functions. Importantly, its deletion causes lethality and defects in DNA segregation. This work defines the dramatic changes in NPC composition during A. nidulans mitosis and provides insight into how NPC disassembly may be integrated with mitosis.
Figures
Figure 1.
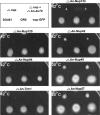
Conditional phenotypes associated with Nup deletions. Strains were spot-inoculated and incubated as indicated. The temperature sensitivity of strains at 42°C (ΔAn-nup120, ΔAn-nup84, ΔAn-trm1, ΔAn-nup133, and ΔAn-nup42) and the cold sensitivity of strains at 20°C (ΔAn-nup49 and ΔAn-nup57) are shown after 2 d (42°C) or 5 d of growth (20°C). As indicated by the key, in each plate the original deletion in the ΔAn-ku70 strain and out-crossed strains have been inoculated. As controls, ΔAn-ku70 and wild-type strains, along with strains carrying the GFP-tagged version of the deleted gene, are also inoculated. All strains grew similarly to wild type at 32°C.
Figure 2.
Gene deletion of An-nup2. (A) Conidia from pyrG+ control or Δ_nup2_::pyrG deletion transformants were replica streaked onto nonselective (+UU) and selective plates (−UU) and grown for 2 d before photography. Conidia that generate colonies on +UU and −UU media are from pyrG+ nonheterokaryon colonies, whereas conidia that form colonies on +UU but not −UU media are from ΔAn-nup2::pyrG/An-nup2 pyrG89 heterokaryons. (B) Diagnostic PCR reactions using DNA isolated from a wild-type strain (Wt.), a putative Δ_nup2_/nup2+ heterokaryon (Het.), and from a colony after streaking conidia from the heterokaryon on nonselective media (Het. +UU). The marker (M) is λ_HindIII_. (C) Wild-type germlings with 2, 4, or 8 nuclei stained with DAPI are shown (a–c). Also shown are Δ_nup2_ germlings (d–h) stained with DAPI indicating miss-segregated DNA (arrows) or a single large nucleus (arrow head). (D) Germlings grown from conidia of the Δ_nup2_/nup2+ heterokaryon on nonselective media for the pyrG89 marker after DAPI staining are shown. In each case to the left are pyrG89 cells able to grow normally on the +UU supplemented media. Cells to the right are Δ_nup2_ cells and show the same phenotypes as seen on selective media (C, e–h) with miss-segregated DNA (arrows) or single large nuclei (arrow head). Scale bars, ∼5 μm.
Figure 3.
Growth characteristics after essential gene deletions. (A) Depicted is a bright field image of conidia from a pyrG89 strain (GR5) grown on selective (−UU) or nonselective media for pyrG89 (+UU) for 1 or 3 d at 22°C. Note the germinating pyrG89 conidia are unable to form a germtube even after 3 d growth on selective media, although they swell considerably. (B) Growth characteristics of conidia from 18 different essential gene deletions derived from heterokaryons containing nuclei with the deleted allele (Δ_gene X_::pyrG+) and pyrG89 nuclei with the wild-type allele (Gene X+; _pyrG_−). Growth was for 1 or 3 d at 22°C on media selective for pyrG+. In each panel the pyrG89 cells are the unpolarized cells, and the null cells are the germlings with varying growth capabilities. Scale bar, ∼20 μm.
Figure 4.
Nuclear defects after nuclear transport gene deletions. Shown are micrographs of representative germlings with the indicated gene deletions stained with DAPI to reveal nuclear number and morphology after 24 h growth at 22°C. Note varying amounts of growth and nuclear morphologies compared with wild-type cells (Figure 2C, a–c). Scale bar, ∼5 μm.
Figure 5.
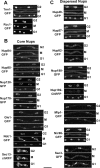
Cell cycle distribution of endogenously tagged Nups during mitosis. (A–C) For each endogenously tagged strain, images are shown from time-course microscopy during mitosis. Cells containing two nuclei, which divide to generate four nuclei, are shown. The images are maximum intensity projections from multiple _Z_-sections. Proteins are tagged either with GFP or with the mCherry variant of mRFP (Shaner et al., 2004), indicated as chRFP. For each tagged strain, cells are shown in late G2 before mitosis (G2), at midmitosis just before nuclear division (M), and in G1 when nuclear division has been completed (G1). For each set of images the original movie file is presented as a supplementary file (An-Trm1-GFP Video 1, An-Rcc1-GFP Video 2, An-Nup84-GFP Video 3, An-Nup85- GFP Video 4, An-Nup120-GFP Video 5, An-Nup170-GFP Video 6, An-Gle1-GFP Video 7, An-Ndc1-GFP Video 8, An-Sec13-chRFP Video 9, An-Nup49-GFP Video 11, An-Nup57-GFP Video 12, An-Nup82-GFP Video 13, An-Nup188-GFP Video 14, An-Nup192-chRFP Video 15, An-Mlp1-GFP Video 16, An-Nic96-GFP Video 17, and An-Sac3-chRFP Video 18). Scale bar, ∼5 μm.
Figure 6.
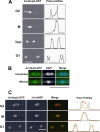
An-Nup2-GFP relocates from the NPC to DNA during mitosis. (A) The GFP signal for An-Nup2-GFP is shown during mitosis along with the pixel profile through the dividing nucleus. The micrographs are maximum intensity projections from multiple _Z_-sections from time-lapse confocal microscopy. The movie file is presented as a supplementary file (Supplementary Video 19). (B) The An-Nup2-GFP signal is shown in fixed interphase and mitotic cells. The location of DNA is shown by DAPI staining, and the merge of the two signals is shown. (C) The An-Nup2-GFP and histone H1-mRFP signals are shown as maximum intensity projections from live cell imaging in G2, mitosis (M), and G1 as indicated. A merged image is shown as are the pixel profiles for the An-Nup2-GFP (green) and histone H1-mRFP (red) signals through the dividing nucleus. The corresponding movie file is presented as a supplementary file (Supplementary Video 20). Scale bars, ∼5 μm.
Figure 7.
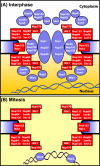
Mitotic changes in the composition of the A. nidulans NPC. (A) Depicted are the predicted location of A. nidulans Nups during interphase and (B) during mitosis. It is noteworthy that all the Nups we have found to disperse from the NPC (in blue) are known to be peripheral Nups, whereas the core Nups (in red) consist of the Nup84 subcomplex, membrane Nups and An-Gle1 and An-Nup170. All of these are considered core Nups in other systems with the exception of An-Gle1, which is a peripheral Nup involved in RNA export. An-Nup2 is unique as it locates from the NPC to chromatin during mitosis. The model is based on data from the current study and De Souza et al. (2004). The figure is modeled after Powers and Dasso (2004).
Similar articles
- The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex.
Liu HL, De Souza CP, Osmani AH, Osmani SA. Liu HL, et al. Mol Biol Cell. 2009 Jan;20(2):616-30. doi: 10.1091/mbc.e08-06-0628. Epub 2008 Nov 19. Mol Biol Cell. 2009. PMID: 19019988 Free PMC article. - The mode of mitosis is dramatically modified by deletion of a single nuclear pore complex gene in Aspergillus nidulans.
Chemudupati M, Johns M, Osmani SA. Chemudupati M, et al. Fungal Genet Biol. 2019 Sep;130:72-81. doi: 10.1016/j.fgb.2019.04.010. Epub 2019 Apr 23. Fungal Genet Biol. 2019. PMID: 31026588 - Mitotic nuclear pore complex segregation involves Nup2 in Aspergillus nidulans.
Suresh S, Markossian S, Osmani AH, Osmani SA. Suresh S, et al. J Cell Biol. 2017 Sep 4;216(9):2813-2826. doi: 10.1083/jcb.201610019. Epub 2017 Jul 26. J Cell Biol. 2017. PMID: 28747316 Free PMC article. - Poring over chromosomes: mitotic nuclear pore complex segregation.
Suresh S, Osmani SA. Suresh S, et al. Curr Opin Cell Biol. 2019 Jun;58:42-49. doi: 10.1016/j.ceb.2019.01.002. Epub 2019 Feb 22. Curr Opin Cell Biol. 2019. PMID: 30798206 Review. - The Role of Nucleoporin Elys in Nuclear Pore Complex Assembly and Regulation of Genome Architecture.
Shevelyov YY. Shevelyov YY. Int J Mol Sci. 2020 Dec 13;21(24):9475. doi: 10.3390/ijms21249475. Int J Mol Sci. 2020. PMID: 33322130 Free PMC article. Review.
Cited by
- Nup2 performs diverse interphase functions in Aspergillus nidulans.
Suresh S, Markossian S, Osmani AH, Osmani SA. Suresh S, et al. Mol Biol Cell. 2018 Dec 15;29(26):3144-3154. doi: 10.1091/mbc.E18-04-0223. Epub 2018 Oct 24. Mol Biol Cell. 2018. PMID: 30355026 Free PMC article. - The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex.
Liu HL, De Souza CP, Osmani AH, Osmani SA. Liu HL, et al. Mol Biol Cell. 2009 Jan;20(2):616-30. doi: 10.1091/mbc.e08-06-0628. Epub 2008 Nov 19. Mol Biol Cell. 2009. PMID: 19019988 Free PMC article. - Nuclear transporters in a multinucleated organism: functional and localization analyses in Aspergillus nidulans.
Markina-Iñarrairaegui A, Etxebeste O, Herrero-García E, Araújo-Bazán L, Fernández-Martínez J, Flores JA, Osmani SA, Espeso EA. Markina-Iñarrairaegui A, et al. Mol Biol Cell. 2011 Oct;22(20):3874-86. doi: 10.1091/mbc.E11-03-0262. Epub 2011 Aug 31. Mol Biol Cell. 2011. PMID: 21880896 Free PMC article. - Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells.
Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Dultz E, et al. J Cell Biol. 2008 Mar 10;180(5):857-65. doi: 10.1083/jcb.200707026. Epub 2008 Mar 3. J Cell Biol. 2008. PMID: 18316408 Free PMC article. - Nuclear translocation of RanGAP1 coincides with virtual nuclear envelope breakdown in fission yeast meiosis.
Asakawa H, Hiraoka Y, Haraguchi T. Asakawa H, et al. Commun Integr Biol. 2011 May;4(3):312-4. doi: 10.4161/cib.4.3.14808. Epub 2011 May 1. Commun Integr Biol. 2011. PMID: 21980566 Free PMC article.
References
- Alcazar-Roman A. R., Tran E. J., Guo S., Wente S. R. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006;8:711–716. - PubMed
- Allen N. P., Patel S. S., Huang L., Chalkley R. J., Burlingame A., Lutzmann M., Hurt E. C., Rexach M. Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell Proteomics. 2002;1:930–946. - PubMed
- Bachewich C., Masker K., Osmani S. The polo-like kinase PLKA is required for initiation and progression through mitosis in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2005;55:572–587. - PubMed
- Bai S. W., Rouquette J., Umeda M., Faigle W., Loew D., Sazer S., Doye V. The fission yeast Nup107–120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell. Biol. 2004;24:6379–6392. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases