Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi - PubMed (original) (raw)
Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi
Lisa A Fitzgerald et al. Virology. 2007.
Abstract
Viruses MT325 and FR483, members of the family Phycodnaviridae, genus Chlorovirus, infect the fresh water, unicellular, eukaryotic, chlorella-like green alga, Chlorella Pbi. The 314,335-bp genome of MT325 and the 321,240-bp genome of FR483 are the first viruses that infect Chlorella Pbi to have their genomes sequenced and annotated. Furthermore, these genomes are the two smallest chlorella virus genomes sequenced to date, MT325 has 331 putative protein-encoding and 10 tRNA-encoding genes and FR483 has 335 putative protein-encoding and 9 tRNA-encoding genes. The protein-encoding genes are almost evenly distributed on both strands, and intergenic space is minimal. Approximately 40% of the viral gene products resemble entries in public databases, including some that are the first of their kind to be detected in a virus. For example, these unique gene products include an aquaglyceroporin in MT325, a potassium ion transporter protein and an alkyl sulfatase in FR483, and a dTDP-glucose pyrophosphorylase in both viruses. Comparison of MT325 and FR483 protein-encoding genes with the prototype chlorella virus PBCV-1 indicates that approximately 82% of the genes are present in all three viruses.
Figures
Fig. 1
Comparison of three sequenced chlorella viruses, PBCV-1, MT325, and FR483, major ORFs with blastp dot plots. The dots represent ORF homology between two viruses with an E-value of less than 0.001.
Fig. 2
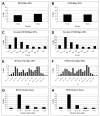
General characteristics of MT325 and FR483 major ORFs. Direction of the MT325 (A) and FR483 (B) ORFs. Size of the major MT325 (C) and FR483 (D) ORFs. Predicted isoelectric points of the major MT325 (E) and FR483 (F) ORFs. Intergenic space between major MT325 (G) and FR483 (H) ORFs.
Fig. 3
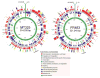
Map of the MT325 and FR483 genomes. The predicted open reading frames are shown in both orientations and colored according to their functional category. The inner green circle represents the percent A+T across the genome using a 25 bp window.
Similar articles
- Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea.
Fitzgerald LA, Graves MV, Li X, Hartigan J, Pfitzner AJ, Hoffart E, Van Etten JL. Fitzgerald LA, et al. Virology. 2007 Jun 5;362(2):350-61. doi: 10.1016/j.virol.2006.12.028. Epub 2007 Feb 5. Virology. 2007. PMID: 17276475 Free PMC article. - Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A.
Fitzgerald LA, Graves MV, Li X, Feldblyum T, Nierman WC, Van Etten JL. Fitzgerald LA, et al. Virology. 2007 Feb 20;358(2):472-84. doi: 10.1016/j.virol.2006.08.033. Epub 2006 Oct 4. Virology. 2007. PMID: 17027058 Free PMC article. - Phycodnaviridae--large DNA algal viruses.
Van Etten JL, Graves MV, Müller DG, Boland W, Delaroque N. Van Etten JL, et al. Arch Virol. 2002 Aug;147(8):1479-516. doi: 10.1007/s00705-002-0822-6. Arch Virol. 2002. PMID: 12181671 Review. - Analysis of 74 kb of DNA located at the right end of the 330-kb chlorella virus PBCV-1 genome.
Li Y, Lu Z, Sun L, Ropp S, Kutish GF, Rock DL, Van Etten JL. Li Y, et al. Virology. 1997 Oct 27;237(2):360-77. doi: 10.1006/viro.1997.8805. Virology. 1997. PMID: 9356347 - Unusual life style of giant chlorella viruses.
Van Etten JL. Van Etten JL. Annu Rev Genet. 2003;37:153-95. doi: 10.1146/annurev.genet.37.110801.143915. Annu Rev Genet. 2003. PMID: 14616059 Review.
Cited by
- Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses.
Jeanniard A, Dunigan DD, Gurnon JR, Agarkova IV, Kang M, Vitek J, Duncan G, McClung OW, Larsen M, Claverie JM, Van Etten JL, Blanc G. Jeanniard A, et al. BMC Genomics. 2013 Mar 8;14:158. doi: 10.1186/1471-2164-14-158. BMC Genomics. 2013. PMID: 23497343 Free PMC article. - Viral metagenomics analysis of planktonic viruses in East Lake, Wuhan, China.
Ge X, Wu Y, Wang M, Wang J, Wu L, Yang X, Zhang Y, Shi Z. Ge X, et al. Virol Sin. 2013 Oct;28(5):280-90. doi: 10.1007/s12250-013-3365-y. Epub 2013 Sep 30. Virol Sin. 2013. PMID: 24132758 Free PMC article. - Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea.
Fitzgerald LA, Graves MV, Li X, Hartigan J, Pfitzner AJ, Hoffart E, Van Etten JL. Fitzgerald LA, et al. Virology. 2007 Jun 5;362(2):350-61. doi: 10.1016/j.virol.2006.12.028. Epub 2007 Feb 5. Virology. 2007. PMID: 17276475 Free PMC article. - Deep RNA sequencing reveals hidden features and dynamics of early gene transcription in Paramecium bursaria chlorella virus 1.
Blanc G, Mozar M, Agarkova IV, Gurnon JR, Yanai-Balser G, Rowe JM, Xia Y, Riethoven JJ, Dunigan DD, Van Etten JL. Blanc G, et al. PLoS One. 2014 Mar 7;9(3):e90989. doi: 10.1371/journal.pone.0090989. eCollection 2014. PLoS One. 2014. PMID: 24608750 Free PMC article. - Genetic diversity and temporal dynamics of phytoplankton viruses in East Lake, China.
Wang MN, Ge XY, Wu YQ, Yang XL, Tan B, Zhang YJ, Shi ZL. Wang MN, et al. Virol Sin. 2015 Aug;30(4):290-300. doi: 10.1007/s12250-015-3603-6. Epub 2015 Aug 5. Virol Sin. 2015. PMID: 26248585 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Bannister JV, Bannister WH, Rotilio G. Aspects of the structure, function, and applications of superoxide dismutase. CRC Rev Biochem. 1987;22:111–180. - PubMed
- Belfield GP, Tuite MF. Translation elongation factor 3: a fungus-specific translation factor? Mol Microbiol. 1993;9:411–418. - PubMed
- Bubeck JA, Pfitzner AJP. Isolation and characterization of a new type of chlorovirus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea. J Gen Virol. 2005;86:2871–2877. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases