Long intervals of stasis punctuated by bursts of positive selection in the seasonal evolution of influenza A virus - PubMed (original) (raw)
Long intervals of stasis punctuated by bursts of positive selection in the seasonal evolution of influenza A virus
Yuri I Wolf et al. Biol Direct. 2006.
Abstract
Background: The interpandemic evolution of the influenza A virus hemagglutinin (HA) protein is commonly considered a paragon of rapid evolutionary change under positive selection in which amino acid replacements are fixed by virtue of their effect on antigenicity, enabling the virus to evade immune surveillance.
Results: We performed phylogenetic analyses of the recently obtained large and relatively unbiased samples of the HA sequences from 1995-2005 isolates of the H3N2 and H1N1 subtypes of influenza A virus. Unexpectedly, it was found that the evolution of H3N2 HA includes long intervals of generally neutral sequence evolution without apparent substantial antigenic change ("stasis" periods) that are characterized by an excess of synonymous over nonsynonymous substitutions per site, lack of association of amino acid replacements with epitope regions, and slow extinction of coexisting virus lineages. These long periods of stasis are punctuated by shorter intervals of rapid evolution under positive selection during which new dominant lineages quickly displace previously coexisting ones. The preponderance of positive selection during intervals of rapid evolution is supported by the dramatic excess of amino acid replacements in the epitope regions of HA compared to replacements in the rest of the HA molecule. In contrast, the stasis intervals showed a much more uniform distribution of replacements over the HA molecule, with a statistically significant difference in the rate of synonymous over nonsynonymous substitution in the epitope regions between the two modes of evolution. A number of parallel amino acid replacements - the same amino acid substitution occurring independently in different lineages - were also detected in H3N2 HA. These parallel mutations were, largely, associated with periods of rapid fitness change, indicating that there are major limitations on evolutionary pathways during antigenic change. The finding that stasis is the prevailing modality of H3N2 evolution suggests that antigenic changes that lead to an increase in fitness typically result from epistatic interactions between several amino acid substitutions in the HA and, perhaps, other viral proteins. The strains that become dominant due to increased fitness emerge from low frequency strains thanks to the last amino acid replacement that completes the set of replacements required to produce a significant antigenic change; no subset of substitutions results in a biologically significant antigenic change and corresponding fitness increase. In contrast to H3N2, no clear intervals of evolution under positive selection were detected for the H1N1 HA during the same time span. Thus, the ascendancy of H1N1 in some seasons is, most likely, caused by the drop in the relative fitness of the previously prevailing H3N2 lineages as the fraction of susceptible hosts decreases during the stasis intervals.
Conclusion: We show that the common view of the evolution of influenza virus as a rapid, positive selection-driven process is, at best, incomplete. Rather, the interpandemic evolution of influenza appears to consist of extended intervals of stasis, which are characterized by neutral sequence evolution, punctuated by shorter intervals of rapid fitness increase when evolutionary change is driven by positive selection. These observations have implications for influenza surveillance and vaccine formulation; in particular, the possibility exists that parallel amino acid replacements could serve as a predictor of new dominant strains.
Reviewers: Ron Fouchier (nominated by Andrey Rzhetsky), David Krakauer, Christopher Lee.
Figures
Figure 1
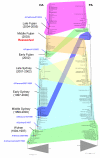
A comparison of the phylogenetic trees for the HA and PA genes of the H3N2 subtype of influenza A virus from 1994–2005. The colored connectors indicate major discrepancies between the gene trees that result from reassortment. The subtrees corresponding to sequences from isolates prior to 1994 are collapsed to focus on isolates from 1994 through 2005 and portions of the trees are labeled ("Wuhan", "Middle Sydney", etc.) with names derived from antigenically distinctive isolates that dominated during particular time intervals. Approximate positions of the vaccine isolates are indicated by red asterisks and blue, italicized labels.
Figure 2
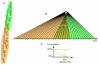
Calculation of extinction times. A. An arbitrary trunk branch (highlighted in red) divides a tree into ancestral (orange shading) and descendant (green shading) parts. B. Terminal nodes (isolates) are arranged along the time axis; the overlap area denotes the period of time during which the descendant lineages drive the ancestral lineages to extinction. C. Extinction curve of the isolates from the ancestral part of the tree, for the overlap interval shown in (B); the 90% extinction point is indicated.
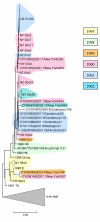
Figure 3
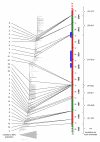
Periods of stasis and rapid fitness change in the evolution of H3N2 HA. Red intervals indicate stasis and green intervals indicate rapid change. Blue bars show the intervals of H1N1 dominance. The numbers to the left of the tree indicate the extinction time of the co-existing descendants of the given node. The ratios to the right are: (nonsynonymous mutations in epitopes + nonsynonymous mutations outside epitopes)/synonymous mutations in the trunk branches.
Figure 4
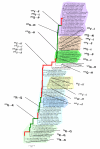
Parallel amino acid replacements in H3N2 HA. The replacements in epitope regions are shown in boldface. Color-code for shading of lineages is as in Figure 1. Stasis intervals are indicated in red in the main trunk of the tree, while intervals associated with rapid fitness change appear in green, as in Figure 2.
Figure 5
Prolonged stasis in the evolution of H1N1 HA. Isolates from the same influenza season are shaded in the same color.
Similar articles
- The evolution of human influenza A viruses from 1999 to 2006: a complete genome study.
Bragstad K, Nielsen LP, Fomsgaard A. Bragstad K, et al. Virol J. 2008 Mar 7;5:40. doi: 10.1186/1743-422X-5-40. Virol J. 2008. PMID: 18325125 Free PMC article. - Evolution of the hemagglutinin expressed by human influenza A(H1N1)pdm09 and A(H3N2) viruses circulating between 2008-2009 and 2013-2014 in Germany.
Wedde M, Biere B, Wolff T, Schweiger B. Wedde M, et al. Int J Med Microbiol. 2015 Oct;305(7):762-75. doi: 10.1016/j.ijmm.2015.08.030. Epub 2015 Aug 21. Int J Med Microbiol. 2015. PMID: 26416089 - Deep Sequencing Reveals Potential Antigenic Variants at Low Frequencies in Influenza A Virus-Infected Humans.
Dinis JM, Florek KR, Fatola OO, Moncla LH, Mutschler JP, Charlier OK, Meece JK, Belongia EA, Friedrich TC. Dinis JM, et al. J Virol. 2016 Jan 6;90(7):3355-65. doi: 10.1128/JVI.03248-15. J Virol. 2016. PMID: 26739054 Free PMC article. - Epidemiological and genetic characterization of pH1N1 and H3N2 influenza viruses circulated in MENA region during 2009-2017.
Al Khatib HA, Al Thani AA, Gallouzi I, Yassine HM. Al Khatib HA, et al. BMC Infect Dis. 2019 Apr 11;19(1):314. doi: 10.1186/s12879-019-3930-6. BMC Infect Dis. 2019. PMID: 30971204 Free PMC article. - Specialized springtail predation by Loricera beetles: An example of evolutionary stasis across the K-Pg extinction.
Li YD, Tihelka E, Engel MS, Huang D, Cai C. Li YD, et al. Innovation (Camb). 2024 Feb 29;5(3):100601. doi: 10.1016/j.xinn.2024.100601. eCollection 2024 May 6. Innovation (Camb). 2024. PMID: 38745760 Free PMC article. Review. No abstract available.
Cited by
- Integrating genotypes and phenotypes improves long-term forecasts of seasonal influenza A/H3N2 evolution.
Huddleston J, Barnes JR, Rowe T, Xu X, Kondor R, Wentworth DE, Whittaker L, Ermetal B, Daniels RS, McCauley JW, Fujisaki S, Nakamura K, Kishida N, Watanabe S, Hasegawa H, Barr I, Subbarao K, Barrat-Charlaix P, Neher RA, Bedford T. Huddleston J, et al. Elife. 2020 Sep 2;9:e60067. doi: 10.7554/eLife.60067. Elife. 2020. PMID: 32876050 Free PMC article. - Waning immunity drives respiratory virus evolution and reinfection.
Bull JJ, Koelle K, Antia R. Bull JJ, et al. bioRxiv [Preprint]. 2024 Aug 8:2024.07.23.604867. doi: 10.1101/2024.07.23.604867. bioRxiv. 2024. PMID: 39091870 Free PMC article. Preprint. - Evolutionary dynamics of N-glycosylation sites of influenza virus hemagglutinin.
Cherry JL, Lipman DJ, Nikolskaya A, Wolf YI. Cherry JL, et al. PLoS Curr. 2009 Aug 18;1:RRN1001. doi: 10.1371/currents.rrn1001. PLoS Curr. 2009. PMID: 20025194 Free PMC article. - Recent human influenza A/H3N2 virus evolution driven by novel selection factors in addition to antigenic drift.
Memoli MJ, Jagger BW, Dugan VG, Qi L, Jackson JP, Taubenberger JK. Memoli MJ, et al. J Infect Dis. 2009 Oct 15;200(8):1232-41. doi: 10.1086/605893. J Infect Dis. 2009. PMID: 19743921 Free PMC article. - Turnover of SARS-CoV-2 Lineages Shaped the Pandemic and Enabled the Emergence of New Variants in the State of Rio de Janeiro, Brazil.
Francisco Junior RDS, Lamarca AP, de Almeida LGP, Cavalcante L, Machado DT, Martins Y, Brustolini O, Gerber AL, Guimarães APC, Gonçalves RB, Alves C, Mariani D, Cruz TF, de Souza IV, de Carvalho EM, Ribeiro MS, Carvalho S, da Silva FD, Garcia MHO, de Souza LM, da Silva CG, Ribeiro CLP, Cavalcanti AC, de Mello CMB, Struchiner CJ, Tanuri A, de Vasconcelos ATR. Francisco Junior RDS, et al. Viruses. 2021 Oct 7;13(10):2013. doi: 10.3390/v13102013. Viruses. 2021. PMID: 34696443 Free PMC article.
References
- CDC Flu Activity, Reports & Surveillance methods in the United States http://www.cdc.gov/flu/weekly/fluactivity.htm
LinkOut - more resources
Full Text Sources
Other Literature Sources