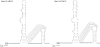Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features - PubMed (original) (raw)
Comparative Study
. 2007 Feb;81(4):1574-85.
doi: 10.1128/JVI.02182-06. Epub 2006 Nov 22.
Ming Wang, Susanna K P Lau, Huifang Xu, Rosana W S Poon, Rongtong Guo, Beatrice H L Wong, Kai Gao, Hoi-Wah Tsoi, Yi Huang, Kenneth S M Li, Carol S F Lam, Kwok-Hung Chan, Bo-Jian Zheng, Kwok-Yung Yuen
Affiliations
- PMID: 17121802
- PMCID: PMC1797546
- DOI: 10.1128/JVI.02182-06
Comparative Study
Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features
Patrick C Y Woo et al. J Virol. 2007 Feb.
Abstract
Twelve complete genomes of three novel coronaviruses-bat coronavirus HKU4 (bat-CoV HKU4), bat-CoV HKU5 (putative group 2c), and bat-CoV HKU9 (putative group 2d)-were sequenced. Comparative genome analysis showed that the various open reading frames (ORFs) of the genomes of the three coronaviruses had significantly higher amino acid identities to those of other group 2 coronaviruses than group 1 and 3 coronaviruses. Phylogenetic trees constructed using chymotrypsin-like protease, RNA-dependent RNA polymerase, helicase, spike, and nucleocapsid all showed that the group 2a and 2b and putative group 2c and 2d coronaviruses are more closely related to each other than to group 1 and 3 coronaviruses. Unique genomic features distinguishing between these four subgroups, including the number of papain-like proteases, the presence or absence of hemagglutinin esterase, small ORFs between the membrane and nucleocapsid genes and ORFs (NS7a and NS7b), bulged stem-loop and pseudoknot structures downstream of the nucleocapsid gene, transcription regulatory sequence, and ribosomal recognition signal for the envelope gene, were also observed. This is the first time that NS7a and NS7b downstream of the nucleocapsid gene has been found in a group 2 coronavirus. The high Ka/Ks ratio of NS7a and NS7b in bat-CoV HKU9 implies that these two group 2d-specific genes are under high selective pressure and hence are rapidly evolving. The four subgroups of group 2 coronaviruses probably originated from a common ancestor. Further molecular epidemiological studies on coronaviruses in the bats of other countries, as well as in other animals, and complete genome sequencing will shed more light on coronavirus diversity and their evolutionary histories.
Figures
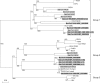
FIG. 1.
Phylogenetic analysis of amino acid sequences of the 393-bp fragment of RNA-dependent RNA polymerase of coronaviruses identified from bats in the present study. The tree was constructed by the neighbor-joining method using the Jukes-Cantor correction and bootstrap values calculated from 1,000 trees. The scale bar indicates the estimated number of substitutions per 50 amino acids. Coronaviruses identified in the present study are shown in boldface. Coronaviruses from bats are shaded in gray. HCoV-229E (NC_002645); PEDV, porcine epidemic diarrhea virus (NC_003436); TGEV(NC_002306); FIPV (AY994055); HCoV-NL63 NL63 (NC_005831); bat-CoV HKU2 (DQ249235), HKU4 (DQ074652), HKU5 (DQ249219), HKU6 (DQ249224), HKU7 (DQ249226), and HKU8 (DQ249228); CoV-HKU1 (NC_006577); HCoV-OC43 (NC_005147); MHV, murine hepatitis virus (NC_006852); BCoV, bovine coronavirus (NC_003045); PHEV, porcine hemagglutinating encephalomyelitis virus (NC_007732); SDAV; SARS-CoV (human), human SARS coronavirus (NC_004718); SARS-CoV (Civet), civet SARS-like coronavirus (AY304488); bat-SARS-CoV HKU3, bat-SARS-like coronavirus HKU3 (DQ022305); IBV, infectious bronchitis virus (NC_001451); TCoV, turkey coronavirus (AF124991); IBV-like, IBV isolated from peafowl (AY641576). Other abbreviations are as defined in the text.
FIG. 2.
Genome organizations of bat-CoV HKU4, bat-CoV HKU5, bat-CoV HKU9, and representative coronaviruses from each group. Papain-like proteases (PL1, PL2, and PL) and the nonstructural proteins are represented by white boxes. Hemagglutinin esterase (HE), spike (S), envelope (E), membrane (M), and nucleocapsid (N) are represented by gray boxes.
FIG. 3.
Multiple alignments of PLpro of SARS-CoV, btCoV/133/05 (NC_008315), bat-CoV HKU4, bat-CoV HKU5, bat-CoV HKU9, and IBV and PL2pro of HCoV-229E, TGEV, HCoV-OC43, and MHV. Amino acids conserved across all coronaviruses are highlighted in black. Amino acids conserved in 60 to 90% of the coronaviruses are highlighted in gray. The conserved Cys and His amino acid residues of the catalytic dyad are marked with an asterisk, the conserved postulated metal-chelating Cys and His residues are marked with a “#” symbol, and the conserved aromatic amino acid immediately downstream of the catalytic Cys is marked with a “+” symbol.
FIG. 4.
Predicted bulged stem-loop and pseudoknot structures downstream of N in genomes of bat-CoV HKU4 and bat-CoV HKU5. Stop codons for the N genes are boxed. Broken lines indicate alternative base pairing.
FIG. 5.
Phylogenetic analysis of chymotrypsin-like protease (3CLpro), RNA-dependent RNA polymerase (Pol), helicase (Hel), spike (S), and nucleocapsid (N) of bat-CoV HKU4, bat-CoV HKU5, and bat-CoV HKU9. The trees were constructed by the neighbor-joining method using the Jukes-Cantor correction and bootstrap values calculated from 1,000 trees. We included 327, 949, 609, 1,661, and 582 amino acid positions in 3CLpro, Pol, helicase, S and N, respectively, in the analysis. The scale bar indicates the estimated number of substitutions per 10 amino acids. Abbreviations are as defined in the text or in the legend to Fig. 1.
Similar articles
- Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus.
Woo PC, Lau SK, Lam CS, Lai KK, Huang Y, Lee P, Luk GS, Dyrting KC, Chan KH, Yuen KY. Woo PC, et al. J Virol. 2009 Jan;83(2):908-17. doi: 10.1128/JVI.01977-08. Epub 2008 Oct 29. J Virol. 2009. PMID: 18971277 Free PMC article. - Coronavirus diversity, phylogeny and interspecies jumping.
Woo PC, Lau SK, Huang Y, Yuen KY. Woo PC, et al. Exp Biol Med (Maywood). 2009 Oct;234(10):1117-27. doi: 10.3181/0903-MR-94. Epub 2009 Jun 22. Exp Biol Med (Maywood). 2009. PMID: 19546349 Review. - Molecular diversity of coronaviruses in bats.
Woo PC, Lau SK, Li KS, Poon RW, Wong BH, Tsoi HW, Yip BC, Huang Y, Chan KH, Yuen KY. Woo PC, et al. Virology. 2006 Jul 20;351(1):180-7. doi: 10.1016/j.virol.2006.02.041. Epub 2006 May 2. Virology. 2006. PMID: 16647731 Free PMC article. - Bat origin of human coronaviruses.
Hu B, Ge X, Wang LF, Shi Z. Hu B, et al. Virol J. 2015 Dec 22;12:221. doi: 10.1186/s12985-015-0422-1. Virol J. 2015. PMID: 26689940 Free PMC article. Review.
Cited by
- 'One Health' for the people of Hong Kong and the world.
Kok KH, Tang HV, Jin DY. Kok KH, et al. Sci China Life Sci. 2016 Oct;59(10):1068-1070. doi: 10.1007/s11427-012-4408-6. Sci China Life Sci. 2016. PMID: 23238747 Free PMC article. No abstract available. - Identification of Potent, Broad-Spectrum Coronavirus Main Protease Inhibitors for Pandemic Preparedness.
Barkan DT, Garland K, Zhang L, Eastman RT, Hesse M, Knapp M, Ornelas E, Tang J, Cortopassi WA, Wang Y, King F, Jia W, Nguyen Z, Frank AO, Chan R, Fang E, Fuller D, Busby S, Carias H, Donahue K, Tandeske L, Diagana TT, Jarrousse N, Moser H, Sarko C, Dovala D, Moquin S, Marx VM. Barkan DT, et al. J Med Chem. 2024 Oct 10;67(19):17454-17471. doi: 10.1021/acs.jmedchem.4c01404. Epub 2024 Sep 27. J Med Chem. 2024. PMID: 39332817 Free PMC article. - A real-time PCR assay for bat SARS-like coronavirus detection and its application to Italian greater horseshoe bat faecal sample surveys.
Balboni A, Gallina L, Palladini A, Prosperi S, Battilani M. Balboni A, et al. ScientificWorldJournal. 2012;2012:989514. doi: 10.1100/2012/989514. Epub 2011 Nov 22. ScientificWorldJournal. 2012. PMID: 22654650 Free PMC article. - Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin.
Falzarano D, de Wit E, Martellaro C, Callison J, Munster VJ, Feldmann H. Falzarano D, et al. Sci Rep. 2013;3:1686. doi: 10.1038/srep01686. Sci Rep. 2013. PMID: 23594967 Free PMC article. - Replication of MERS and SARS coronaviruses in bat cells offers insights to their ancestral origins.
Lau SKP, Fan RYY, Luk HKH, Zhu L, Fung J, Li KSM, Wong EYM, Ahmed SS, Chan JFW, Kok RKH, Chan KH, Wernery U, Yuen KY, Woo PCY. Lau SKP, et al. Emerg Microbes Infect. 2018 Dec 10;7(1):209. doi: 10.1038/s41426-018-0208-9. Emerg Microbes Infect. 2018. PMID: 30531999 Free PMC article.
References
- Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. Lopez, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, F. Servant, C. J. Sigrist, and E. M. Zdobnov. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37-40. - PMC - PubMed
- Eickmann, M., S. Becker, H. D. Klenk, H. W. Doerr, K. Stadler, S. Censini, S. Guidotti, V. Masignani, M. Scarselli, M. Mora, C. Donati, J. H. Han, H. C. Song, S. Abrignani, A. Covacci, and R. Rappuoli. 2003. Phylogeny of the SARS coronavirus. Science 302:1504-1505. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources