Long-range cooperative interactions modulate dimerization in SARS 3CLpro - PubMed (original) (raw)
Long-range cooperative interactions modulate dimerization in SARS 3CLpro
Jennifer Barrila et al. Biochemistry. 2006.
Abstract
Severe acute respiratory syndrome (SARS) is an infectious disease caused by the human coronavirus, SARS-CoV. The main viral protease, SARS 3CLpro, is a validated target for the development of antiviral therapies. Since the enzyme is a homodimer and the individual monomers are inactive, two approaches are being used to develop inhibitors: enzyme activity inhibitors that target the active site and dimerization inhibitors. Dimerization inhibitors are usually targeted to the dimerization interface and need to compete with the attractive forces between subunits to be effective. In this paper, we show that the dimerization of SARS 3CLpro is also under allosteric control and that additional and energetically more favorable target sites away from the dimerization interface may also lead to subunit dissociation. We previously identified a cluster of conserved serine residues (Ser139, Ser144, and Ser147) located adjacent to the active site of 3CLpro that could effectively be targeted to inactivate the protease [Bacha, U et al. (2004) Biochemistry 43, 4906-4912]. Mutation of any of these serine residues to alanine had a debilitating effect on the catalytic activity of 3CLpro. In particular, the mutation of Ser147, which does not make any contact with the opposing subunit and is located approximately 9 A away from the dimer interface, totally inhibited dimerization and resulted in a complete loss of enzymatic activity. The finding that residues away from the dimer interface are able to control dimerization defines alternative targets for the design of dimerization inhibitors.
Figures
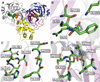
Figure 1
Structure of the SARS 3CLpro dimer at pH 7.6 [pdb file 1UK3 (18)]. (A) Wild type SARS 3CLpro displayed in ribbon representation. Domain 1 (residues 1-100) is shown in blue, domain 2 (residues 101-183) are colored purple, the loop connecting these two domains to domain 3 is colored red and domain 3 (residues 201-306) is shown in yellow. The catalytic residues, His41 and Cys145, are located in the cleft between the first two domains and are displayed as green spheres. The cluster of serines, Ser139, Ser144 and Ser147 are displayed as orange spheres. Important residues surrounding (B) S139A, (C) S144A and (D) S147A are shown.
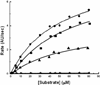
Figure 2
The enzymatic activities of wild type 3CLpro (●), S139A (■), S144A (▲), and S147A (♦) were determined at 25°C with an enzyme concentration of 1 μM in 10mM NaCl, 10mM NaPi, 1mM EDTA, 1mM TCEP (pH 7.4). The initial velocity, which was measured as arbitrary fluorescence units per second, is plotted as a function of substrate concentration.
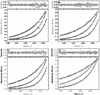
Figure 3
The catalytic efficiencies (_k_cat/_K_m) of (A) wild type 3CLpro, (B) S139A, (C) S144A and (D) S147A are plotted as a function of increasing protease concentration. The _k_cat and _k_m values were obtained by global non-linear least square fit of the initial rate measurements at increasing substrate concentrations. The experiments were performed with in experimental conditions similar to that shown in Fig.2.
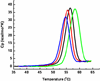
Figure 4
Thermal denaturation of WT (black), S139A (red), S144A (green) and S147A (blue) 3CLpro as determined by differential scanning calorimetry. Excess heat capacity is plotted as a function of temperature. Calorimetric scans for both proteins were performed at identical concentrations (0.2 mg/mL) at a scanning rate of 1°C per minute.
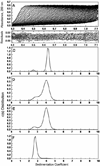
Figure 5
Sedimentation velocity ultracentrifugation of wild type 3CLpro and the serine mutants. The sedimentation of the four recombinant proteins was carried out with a Beckman Coulter XL-I analytical ultracentrifuge at 20°C and 50,000 rpm at concentrations between 0.25 mg/ml and 1 mg/ml. (A) Sedimentation velocity absorbance trace of WT 3CLpro at 280 nm. (B) Residuals of the experimental fit of WT 3CLpro at 0.5 mg/ml (14.8 μM). Continuous sedimentation coefficient distributions at 0.5 mg/ml of (C) WT 3CLpro, (D) S139A, (E) S144A and (F) S147A are shown.
Figure 6
Sedimentation equilibrium ultracentrifugation was carried out and globally analyzed with wild type 3CLpro and the three serine mutants at concentrations between 0.1 and 2 mg/ml (between 2.9 and 59.1 μM). The data sets at 0.5 mg/ml (14.8 μM) are shown for (A) wild type 3CLpro, (B) S139A, (C) S144A and (D) S147A. The lower graphs in each panel show the raw data at 15,000 rpm (Δ), 20,000 rpm (○), and 25,000 rpm (□) and the corresponding global fits displayed in a continuous line. The upper graphs in each panel show the residuals for the given fits.
Similar articles
- Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3C-like protease.
Shi J, Sivaraman J, Song J. Shi J, et al. J Virol. 2008 May;82(9):4620-9. doi: 10.1128/JVI.02680-07. Epub 2008 Feb 27. J Virol. 2008. PMID: 18305031 Free PMC article. - Two adjacent mutations on the dimer interface of SARS coronavirus 3C-like protease cause different conformational changes in crystal structure.
Hu T, Zhang Y, Li L, Wang K, Chen S, Chen J, Ding J, Jiang H, Shen X. Hu T, et al. Virology. 2009 Jun 5;388(2):324-34. doi: 10.1016/j.virol.2009.03.034. Epub 2009 May 5. Virology. 2009. PMID: 19409595 Free PMC article. - The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds.
Báez-Santos YM, St John SE, Mesecar AD. Báez-Santos YM, et al. Antiviral Res. 2015 Mar;115:21-38. doi: 10.1016/j.antiviral.2014.12.015. Epub 2014 Dec 29. Antiviral Res. 2015. PMID: 25554382 Free PMC article. Review. - Activation and maturation of SARS-CoV main protease.
Xia B, Kang X. Xia B, et al. Protein Cell. 2011 Apr;2(4):282-90. doi: 10.1007/s13238-011-1034-1. Epub 2011 Apr 28. Protein Cell. 2011. PMID: 21533772 Free PMC article. Review.
Cited by
- Revealing SARS-CoV-2 Mpro mutation cold and hot spots: Dynamic residue network analysis meets machine learning.
Barozi V, Chakraborty S, Govender S, Morgan E, Ramahala R, Graham SC, Bishop NT, Tastan Bishop Ö. Barozi V, et al. Comput Struct Biotechnol J. 2024 Oct 22;23:3800-3816. doi: 10.1016/j.csbj.2024.10.031. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 39525081 Free PMC article. - An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Westberg M, Su Y, Zou X, Huang P, Rustagi A, Garhyan J, Patel PB, Fernandez D, Wu Y, Hao C, Lo CW, Karim M, Ning L, Beck A, Saenkham-Huntsinger P, Tat V, Drelich A, Peng BH, Einav S, Tseng CK, Blish C, Lin MZ. Westberg M, et al. Sci Transl Med. 2024 Mar 13;16(738):eadi0979. doi: 10.1126/scitranslmed.adi0979. Epub 2024 Mar 13. Sci Transl Med. 2024. PMID: 38478629 - The SARS-CoV-2 Mpro Dimer-Based Screening System: A Synthetic Biology Tool for Identifying Compounds with Dimerization Inhibitory Potential.
Giri-Rachman EA, Effendy VV, Azmi MHS, Yamahoki N, Stephanie R, Agustiyanti DF, Wisnuwardhani PH, Angelina M, Rubiyana Y, Aditama R, Ningrum RA, Wardiana A, Fibriani A. Giri-Rachman EA, et al. ACS Synth Biol. 2024 Feb 16;13(2):509-520. doi: 10.1021/acssynbio.3c00446. Epub 2024 Feb 5. ACS Synth Biol. 2024. PMID: 38316139 Free PMC article. - Systematic Analyses of the Resistance Potential of Drugs Targeting SARS-CoV-2 Main Protease.
Flynn JM, Huang QYJ, Zvornicanin SN, Schneider-Nachum G, Shaqra AM, Yilmaz NK, Moquin SA, Dovala D, Schiffer CA, Bolon DNA. Flynn JM, et al. ACS Infect Dis. 2023 Jul 14;9(7):1372-1386. doi: 10.1021/acsinfecdis.3c00125. Epub 2023 Jun 30. ACS Infect Dis. 2023. PMID: 37390404 Free PMC article. - Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors.
Moghadasi SA, Heilmann E, Khalil AM, Nnabuife C, Kearns FL, Ye C, Moraes SN, Costacurta F, Esler MA, Aihara H, von Laer D, Martinez-Sobrido L, Palzkill T, Amaro RE, Harris RS. Moghadasi SA, et al. Sci Adv. 2023 Mar 29;9(13):eade8778. doi: 10.1126/sciadv.ade8778. Epub 2023 Mar 29. Sci Adv. 2023. PMID: 36989354 Free PMC article.
References
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling A-E, Humphrey CD, Shieh W-J, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang J-Y, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
- Bacha U, Barrila J, Velazquez-Campoy A, Leavitt SA, Freire E. Identification of Novel Inhibitors of the SARS Coronavirus Main Protease 3CLpro. Biochemistry. 2004;43:4906–4912. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous