MetaQTL: a package of new computational methods for the meta-analysis of QTL mapping experiments - PubMed (original) (raw)
MetaQTL: a package of new computational methods for the meta-analysis of QTL mapping experiments
Jean-Baptiste Veyrieras et al. BMC Bioinformatics. 2007.
Abstract
Background: Integration of multiple results from Quantitative Trait Loci (QTL) studies is a key point to understand the genetic determinism of complex traits. Up to now many efforts have been made by public database developers to facilitate the storage, compilation and visualization of multiple QTL mapping experiment results. However, studying the congruency between these results still remains a complex task. Presently, the few computational and statistical frameworks to do so are mainly based on empirical methods (e.g. consensus genetic maps are generally built by iterative projection).
Results: In this article, we present a new computational and statistical package, called MetaQTL, for carrying out whole-genome meta-analysis of QTL mapping experiments. Contrary to existing methods, MetaQTL offers a complete statistical process to establish a consensus model for both the marker and the QTL positions on the whole genome. First, MetaQTL implements a new statistical approach to merge multiple distinct genetic maps into a single consensus map which is optimal in terms of weighted least squares and can be used to investigate recombination rate heterogeneity between studies. Secondly, assuming that QTL can be projected on the consensus map, MetaQTL offers a new clustering approach based on a Gaussian mixture model to decide how many QTL underly the distribution of the observed QTL.
Conclusion: We demonstrate using simulations that the usual model choice criteria from mixture model literature perform relatively well in this context. As expected, simulations also show that this new clustering algorithm leads to a reduction in the length of the confidence interval of QTL location provided that across studies there are enough observed QTL for each underlying true QTL location. The usefulness of our approach is illustrated on published QTL detection results of flowering time in maize. Finally, MetaQTL is freely available at http://bioinformatics.org/mqtl.
Figures
Figure 1
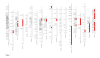
Comparison of model choice criteria: a simulation study. Simulation results for different values of the true number of QTL, K, and the number of observed QTL, q. The vertical bars indicate the probability that the best model selected by the criterion is the true model. The open circles, respectively the dotted lines, represent the mean, respectively the 0.1% and 0.9% quantiles, of the ratios MSEP(2)/MSEP(1) for each criterion.
Figure 2
Performance of AIC: a simulation study. Behavior of the AIC criterion for the 4 distance constraints, _δ_min = 1, 2, 3, 4, and for different values of the true number of QTL, K, and the number of observed QTL, q. The vertical bars indicate the probability than the AIC criterion has selected the true model. The open circles, respectively the dotted lines, represent the mean, respectively the 0.1% and 0.9% quantiles, of the ratios MSEP(2)/MSEP(1).
Figure 3
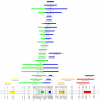
Overview of the maize chromosomes 8 together with the consensus chromosome. Overview of chromosome 8 for the 18 mapping experiments involved in the meta-analysis of flowering time in maize. The first chromosome at the left represents the consensus chromosome obtained by applying the WLS approach as described in the first section of the article (implemented into ConsMap). The filled marker intervals indicate that the standardized residual between the interval distance estimates of the original chromosome and the consensus one exceeded the double-sided 95% percentile of a normalized centered gaussian. This figure has been created by the program MMapView.
Figure 4
Visualization of the QTL meta-analysis result on maize chromosome 8. Result of the meta-analysis of the 34 QTL projected on the consensus chromosome 8. The CI of the meta-QTL positions are indicated on the chromosome by the filled colored areas. The observed QTL positions are depicted by their most probable position (triangle) and CI (segment). Membership probabilities of each initial QTL with respect to meta-QTL is visualized by the proportions of corresponding colored segments. This figure has been created by using MQTLView.
Similar articles
- BioMercator: integrating genetic maps and QTL towards discovery of candidate genes.
Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J. Arcade A, et al. Bioinformatics. 2004 Sep 22;20(14):2324-6. doi: 10.1093/bioinformatics/bth230. Epub 2004 Apr 1. Bioinformatics. 2004. PMID: 15059820 - Meta-analysis of QTL involved in silage quality of maize and comparison with the position of candidate genes.
Truntzler M, Barrière Y, Sawkins MC, Lespinasse D, Betran J, Charcosset A, Moreau L. Truntzler M, et al. Theor Appl Genet. 2010 Nov;121(8):1465-82. doi: 10.1007/s00122-010-1402-x. Epub 2010 Jul 25. Theor Appl Genet. 2010. PMID: 20658277 - MCQTL: multi-allelic QTL mapping in multi-cross design.
Jourjon MF, Jasson S, Marcel J, Ngom B, Mangin B. Jourjon MF, et al. Bioinformatics. 2005 Jan 1;21(1):128-30. doi: 10.1093/bioinformatics/bth481. Epub 2004 Aug 19. Bioinformatics. 2005. PMID: 15319261 - Simultaneous search for multiple QTL using the global optimization algorithm DIRECT.
Ljungberg K, Holmgren S, Carlborg O. Ljungberg K, et al. Bioinformatics. 2004 Aug 12;20(12):1887-95. doi: 10.1093/bioinformatics/bth175. Epub 2004 Mar 25. Bioinformatics. 2004. PMID: 15044246 - Quantitative trait loci mapping in dairy cattle: review and meta-analysis.
Khatkar MS, Thomson PC, Tammen I, Raadsma HW. Khatkar MS, et al. Genet Sel Evol. 2004 Mar-Apr;36(2):163-90. doi: 10.1186/1297-9686-36-2-163. Genet Sel Evol. 2004. PMID: 15040897 Free PMC article. Review.
Cited by
- Genome-wide meta-analysis of QTL for morphological related traits of flag leaf in bread wheat.
Du B, Wu J, Islam MS, Sun C, Lu B, Wei P, Liu D, Chen C. Du B, et al. PLoS One. 2022 Oct 24;17(10):e0276602. doi: 10.1371/journal.pone.0276602. eCollection 2022. PLoS One. 2022. PMID: 36279291 Free PMC article. - Quantitative trait loci identification and meta-analysis for rice panicle-related traits.
Wu Y, Huang M, Tao X, Guo T, Chen Z, Xiao W. Wu Y, et al. Mol Genet Genomics. 2016 Oct;291(5):1927-40. doi: 10.1007/s00438-016-1227-7. Epub 2016 Jul 5. Mol Genet Genomics. 2016. PMID: 27380139 - A meta-quantitative trait loci analysis identified consensus genomic regions and candidate genes associated with grain yield in rice.
Aloryi KD, Okpala NE, Amo A, Bello SF, Akaba S, Tian X. Aloryi KD, et al. Front Plant Sci. 2022 Nov 16;13:1035851. doi: 10.3389/fpls.2022.1035851. eCollection 2022. Front Plant Sci. 2022. PMID: 36466247 Free PMC article. - QTL meta-analysis provides a comprehensive view of loci controlling partial resistance to Aphanomyces euteiches in four sources of resistance in pea.
Hamon C, Coyne CJ, McGee RJ, Lesné A, Esnault R, Mangin P, Hervé M, Le Goff I, Deniot G, Roux-Duparque M, Morin G, McPhee KE, Delourme R, Baranger A, Pilet-Nayel ML. Hamon C, et al. BMC Plant Biol. 2013 Mar 16;13:45. doi: 10.1186/1471-2229-13-45. BMC Plant Biol. 2013. PMID: 23497245 Free PMC article. - Understanding complex genetic architecture of rice grain weight through QTL-meta analysis and candidate gene identification.
Anilkumar C, Sah RP, Muhammed Azharudheen TP, Behera S, Singh N, Prakash NR, Sunitha NC, Devanna BN, Marndi BC, Patra BC, Nair SK. Anilkumar C, et al. Sci Rep. 2022 Aug 16;12(1):13832. doi: 10.1038/s41598-022-17402-w. Sci Rep. 2022. PMID: 35974066 Free PMC article.
References
- Kearsey M, Pooni HS. The genetical analysis of quantitative traits. London: Chapman and Hall; 1996.
- Beavis W. Proceedings of the Forty-Ninth Annual Corn and Sorghum Industry Research Conference. Washington, DC: American Seed Trade Association; 1994. The power and deceit of QTL experiments: lessons from comparative QTL studies; pp. 250–266.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials