Using genomic data to unravel the root of the placental mammal phylogeny - PubMed (original) (raw)
Using genomic data to unravel the root of the placental mammal phylogeny
William J Murphy et al. Genome Res. 2007 Apr.
Abstract
The phylogeny of placental mammals is a critical framework for choosing future genome sequencing targets and for resolving the ancestral mammalian genome at the nucleotide level. Despite considerable recent progress defining superordinal relationships, several branches remain poorly resolved, including the root of the placental tree. Here we analyzed the genome sequence assemblies of human, armadillo, elephant, and opossum to identify informative coding indels that would serve as rare genomic changes to infer early events in placental mammal phylogeny. We also expanded our species sampling by including sequence data from >30 ongoing genome projects, followed by PCR and sequencing validation of each indel in additional taxa. Our data provide support for a sister-group relationship between Afrotheria and Xenarthra (the Atlantogenata hypothesis), which is in turn the sister-taxon to Boreoeutheria. We failed to recover any indels in support of a basal position for Xenarthra (Epitheria), which is suggested by morphology and a recent retroposon analysis, or a hypothesis with Afrotheria basal (Exafricoplacentalia), which is favored by phylogenetic analysis of large nuclear gene data sets. In addition, we identified two retroposon insertions that also support Atlantogenata and none for the alternative hypotheses. A revised molecular timescale based on these phylogenetic inferences suggests Afrotheria and Xenarthra diverged from other placental mammals approximately 103 (95-114) million years ago. We discuss the impacts of this topology on earlier phylogenetic reconstructions and repeat-based inferences of phylogeny.
Figures
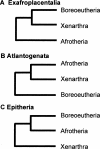
Figure 1.
Three hypotheses for the basal relationships among placental mammal superordinal clades. The name for each hypothesis refers to the ingroup taxa.
Figure 2.
Alignment of PTPRB exon 9 sequences from 49 vertebrate species. A single amino acid deletion shared by all xenarthrans and afrotherians is highlighted with a box. Full-length amino acid sequences are derived from draft genome sequences or traces. Sequences with truncated ends were generated by PCR using conserved primers in the exon flanking the indel. Asterisks indicate gaps in the sequence assemblies or traces.
Figure 3.
Alignment of ZNF367 exon 5 sequences from 46 mammals. A single amino acid insertion shared by all xenarthrans and afrotherians is highlighted with a box. Full-length amino acid sequences are derived from full genome sequences or traces. Sequences with truncated ends were generated by PCR using conserved primers in the exon flanking the indel.
Figure 4.
Alignment of C14orf121 exon 35 sequences from 34 mammals (upper panel), and LAMC2 exon 13 sequences from 28 mammals (lower panel). The indels shared by xenarthrans and afrotherians are highlighted with boxes. Full-length amino acid sequences are derived from draft genome sequences or traces. Sequences with truncated ends were generated by PCR using conserved primers in the exon flanking the indel.
Figure 5.
(A) L1MB5 insertion in an intron of the OTC gene. The upper gel image shows PCR products obtained by amplifying different mammalian genomic DNAs with conserved primers flanking the retroposon insertion site. Below is a partial alignment of the exonic (underlined)/intronic sequences flanking the L1MB5 insertion from 26 placental mammals. The central portion of the inserted sequence has been edited, as shown by the double slash marks. (B) A multispecies alignment showing the second L1MB5 insertion in an intron of the RABGAP1L gene from 23 placental mammals. Direct repeats are highlighted in gray.
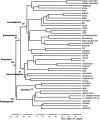
Figure 6.
A molecular timescale for placental mammals based on the data set from Roca et al. (2004), 13 fossil constraints (Springer et al. 2003), and a mean prior of 105 Mya for the placental root. Divergence estimates are shown for several key superordinal clades (for a full list of divergence times and confidence intervals, see Supplemental Table 2).
Similar articles
- Molecular phylogeny of living xenarthrans and the impact of character and taxon sampling on the placental tree rooting.
Delsuc F, Scally M, Madsen O, Stanhope MJ, de Jong WW, Catzeflis FM, Springer MS, Douzery EJ. Delsuc F, et al. Mol Biol Evol. 2002 Oct;19(10):1656-71. doi: 10.1093/oxfordjournals.molbev.a003989. Mol Biol Evol. 2002. PMID: 12270893 - A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships.
Amrine-Madsen H, Koepfli KP, Wayne RK, Springer MS. Amrine-Madsen H, et al. Mol Phylogenet Evol. 2003 Aug;28(2):225-40. doi: 10.1016/s1055-7903(03)00118-0. Mol Phylogenet Evol. 2003. PMID: 12878460 - Early history of mammals is elucidated with the ENCODE multiple species sequencing data.
Nikolaev S, Montoya-Burgos JI, Margulies EH; NISC Comparative Sequencing Program; Rougemont J, Nyffeler B, Antonarakis SE. Nikolaev S, et al. PLoS Genet. 2007 Jan 5;3(1):e2. doi: 10.1371/journal.pgen.0030002. PLoS Genet. 2007. PMID: 17206863 Free PMC article. - Mammal madness: is the mammal tree of life not yet resolved?
Foley NM, Springer MS, Teeling EC. Foley NM, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Jul 19;371(1699):20150140. doi: 10.1098/rstb.2015.0140. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27325836 Free PMC article. Review. - The chromosomes of Afrotheria and their bearing on mammalian genome evolution.
Svartman M, Stanyon R. Svartman M, et al. Cytogenet Genome Res. 2012;137(2-4):144-53. doi: 10.1159/000341387. Epub 2012 Aug 3. Cytogenet Genome Res. 2012. PMID: 22868637 Review.
Cited by
- Femora nutrient foramina and aerobic capacity in giant extinct xenarthrans.
Varela L, Tambusso S, Fariña R. Varela L, et al. PeerJ. 2024 Aug 7;12:e17815. doi: 10.7717/peerj.17815. eCollection 2024. PeerJ. 2024. PMID: 39131616 Free PMC article. - Accurate, scalable, and fully automated inference of species trees from raw genome assemblies using ROADIES.
Gupta A, Mirarab S, Turakhia Y. Gupta A, et al. bioRxiv [Preprint]. 2024 Jun 1:2024.05.27.596098. doi: 10.1101/2024.05.27.596098. bioRxiv. 2024. PMID: 38854139 Free PMC article. Preprint. - Unraveling the role of Xist in X chromosome inactivation: insights from rabbit model and deletion analysis of exons and repeat A.
Liang M, Zhang L, Lai L, Li Z. Liang M, et al. Cell Mol Life Sci. 2024 Mar 29;81(1):156. doi: 10.1007/s00018-024-05151-0. Cell Mol Life Sci. 2024. PMID: 38551746 Free PMC article. - Phylogenomics reveals an almost perfect polytomy among the almost ungulates (Paenungulata).
Bowman J, Enard D, Lynch VJ. Bowman J, et al. bioRxiv [Preprint]. 2023 Dec 8:2023.12.07.570590. doi: 10.1101/2023.12.07.570590. bioRxiv. 2023. PMID: 38106080 Free PMC article. Preprint. - Identification of 3 novel herpesviruses in prosimians with lymphoproliferative disease.
Kunze PE, Cortés-Hinojosa G, Williams CV, Archer LL, Ferrante JA, Wellehan JFX Jr. Kunze PE, et al. J Vet Diagn Invest. 2023 Sep;35(5):514-520. doi: 10.1177/10406387231183431. Epub 2023 Jun 29. J Vet Diagn Invest. 2023. PMID: 37381927 Free PMC article.
References
- Amrine-Madsen H., Koepfli K.P., Wayne R.K., Springer M.S., Koepfli K.P., Wayne R.K., Springer M.S., Wayne R.K., Springer M.S., Springer M.S. A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships. Mol. Phylogenet. Evol. 2003;28:225–240. - PubMed
- Cantrell M.A., Filanoski B.J., Ingermann A.R., Olsson K., DiLuglio N., Lister Z., Wichman H.A., Filanoski B.J., Ingermann A.R., Olsson K., DiLuglio N., Lister Z., Wichman H.A., Ingermann A.R., Olsson K., DiLuglio N., Lister Z., Wichman H.A., Olsson K., DiLuglio N., Lister Z., Wichman H.A., DiLuglio N., Lister Z., Wichman H.A., Lister Z., Wichman H.A., Wichman H.A. An ancient retrovirus-like element contains hotspots for SINE insertion. Genetics. 2001;158:769–777. - PMC - PubMed
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D., Gibson T.J., Higgins D.G., Thompson J.D., Higgins D.G., Thompson J.D., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources