mir-29 regulates Mcl-1 protein expression and apoptosis - PubMed (original) (raw)
mir-29 regulates Mcl-1 protein expression and apoptosis
J L Mott et al. Oncogene. 2007.
Abstract
Cellular expression of Mcl-1, an anti-apoptotic Bcl-2 family member, is tightly regulated. Recently, Bcl-2 expression was shown to be regulated by microRNAs, small endogenous RNA molecules that regulate protein expression through sequence-specific interaction with messenger RNA. By analogy, we reasoned that Mcl-1 expression may also be regulated by microRNAs. We chose human immortalized, but non-malignant, H69 cholangiocyte and malignant KMCH cholangiocarcinoma cell lines for these studies, because Mcl-1 is dysregulated in cells with the malignant phenotype. By in silico analysis, we identified a putative target site in the Mcl-1 mRNA for the mir-29 family, and found that mir-29b was highly expressed in cholangiocytes. Interestingly, mir-29b was downregulated in malignant cells, consistent with Mcl-1 protein upregulation. Enforced mir-29b expression reduced Mcl-1 protein expression in KMCH cells. This effect was direct, as mir-29b negatively regulated the expression of an Mcl-1 3' untranslated region (UTR)-based reporter construct. Enforced mir-29b expression reduced Mcl-1 cellular protein levels and sensitized the cancer cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) cytotoxicity. Transfection of non-malignant cells (that express high levels of mir-29) with a locked-nucleic acid antagonist of mir-29b increased Mcl-1 levels and reduced TRAIL-mediated apoptosis. Thus mir-29 is an endogenous regulator of Mcl-1 protein expression, and thereby, apoptosis.
Figures
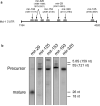
Figure 1. Putative microRNA binding sites on Mcl-1 messenger RNA
Panel a: Schematic of the Mcl-1 3’UTR (#NM_021960) nucleotides 1184−4020. Five microRNAs were predicted to bind and the position of binding to the Mcl-1 transcript is shown in parentheses. Panel b: Northern blot of the five putative Mcl-1 binding microRNAs. RNA enriched for small RNA (<200nt's) from H69 cells was separated and probed for the indicated microRNAs. Note the robust expression of mir-29. Size markers are from SybrGreen-stained gels prior to transfer; 5.8S and 5S are endogenous rRNAs, 26 nt and 18 nt refer to synthetic oligodeoxynucleotides run in an adjacent lane.
Figure 2. Reciprocal expression of mir-29b and Mcl-1
Panel a: Total cellular protein from immortalized non-malignant H69 cells and malignant KMCH cells was probed using a Mcl-1 antibody. Actin was used as a loading control. Panel b: Northern blot for mir-29b using RNA enriched for small RNAs from H69 and KMCH cells. Note that both precursor and mature mir-29b are decreased in KMCH cells. As a loading control, an RNA probe against the small housekeeping RNA U6 (106 nt) was used. Panel c: Quantitative RT-PCR for mir-29b using total RNA from H69 and KMCH cells. Results expressed as copies of mir-29b per copy of Z30 RNA (mean +/− SEM; p < 0.05).
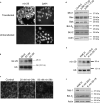
Figure 3. Mcl-1 protein level is negatively regulated by mir-29b
Panel a: Fluorescently-labeled pre-mir-29b was generated by in vitro transcription (Supplemental Methods) and transfected into KMCH cells. After 24 hours, cells were counter-stained with the nuclear stain DAPI, and photographed. For comparison, untransfected cells are shown. Panel b: Northern blot using small RNA from KMCH cells transfected with 20 or 50 nM precursor mir-29b (not fluorescently-tagged) compared to mock-transfected cells. The mature, processed form (23 nt) is shown, demonstrating that the transfected RNA is processed. U6 was probed as a loading control. Panel c: Mcl-1 immunoreactivity in KMCH cells transfected with precursor mir-29b. A Texas red-conjugated secondary antibody was used, and cells were photographed under confocal microscopy. Panel d: Western blots of total cellular protein from KMCH cells transfected with a mir-29b expression vector (p29b). Control cells were transfected with 1 μg of pCDNA, experimentals were transfected with 0.5 or 1 μg p29b. Actin was used as a loading control. Panel e: Northern blot on total RNA isolated from KMCH cells stably transfected with p29b. Two lines were generated, K-mir-29−6 and K-mir-29−19. The housekeeping RNA U6 was probed as a loading control. Panel f: Western blot of total protein from KMCH, K-mir-29−6, and K-mir-29−19 (stably overexpress mir-29b). Actin was used as a loading control.
Figure 4. Effect of the putative mir-29 binding site derived from the Mcl-1 3’UTR on luciferase expression
Panel a: Alignment of mir-29b with the insert derived from the Mcl-1 3’UTR. Note the complementarity at the 5’ end of mir-29b, where the crucial seed region is located. A single-base mutant insert was also synthesized, as shown. Inserts were cloned into the 3’ UTR of the p-Mir-Report vector. Panel b: Luciferase activity in HeLa cells transiently transfected with the luciferase construct alone, or cotransfected with an expression plasmid for mir-29b (p29b). Luciferase vectors were parental (pLuc), luciferase with the Mcl-1-derived 3’UTR insert (pLuc-Mcl-1 3’UTR), or luciferase with the mutated insert (pLuc-Mutant 3’UTR). Mean ± SEM, *p<0.01.
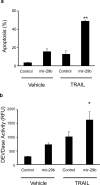
Figure 5. Sensitivity to apoptosis is increased after transfection of mir-29b
Panel a: KMCH cells were transfected with pre-mir-29b RNA or control RNA of the same length (antisense) at 50 nM. After 22 hours, TRAIL was added where indicated at 2 ng/mL in fresh media and the cells were incubated for 4 hours. Cells were then stained with DAPI and cells with apoptotic morphology were counted. Mean ± SEM, **p<0.001. Panel b: In parallel, cells were transfected and treated with TRAIL as in panel a, but after 4 hours caspase 3/7-like (DEVDase) activity was measured. Mean ± SEM, *p<0.01.
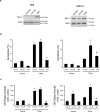
Figure 6. Antagonism of mir-29b protects cells from apoptosis
Panel a: Western blot of total protein isolated from non-malignant H69 cells and malignant KMCH cells transfected with control RNA, pre-mir-29b, or the mir-29 antagonist oligonucleotide, LNA (50nM each). Panel b. Apoptotic morphology measured as in Figure 5 on H69 and KMCH cells transfected as above. Note that H69 cells are sensitive to TRAIL killing, as noted in previous studies, while KMCH cells are relatively resistant unless mir-29b is transfected. Mean ± SEM, **p<0.0001 compared to vehicle treated control; # p<0.01 compared to TRAIL-treated mir-29; ## p<0.0001 compared to TRAIL-treated mir-29. Panel c: Caspase 3/7-like activity (DEVDase) on cells transfected as above. Caspase activity parallels the apoptosis counts using morphology. Mean ± SEM, *p<0.01 compared to vehicle treated control; **p<0.0001 compared to vehicle treated control; ## p<0.0001 compared to TRAIL-treated mir-29.
Similar articles
- Mcl-1 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in human cholangiocarcinoma cells.
Taniai M, Grambihler A, Higuchi H, Werneburg N, Bronk SF, Farrugia DJ, Kaufmann SH, Gores GJ. Taniai M, et al. Cancer Res. 2004 May 15;64(10):3517-24. doi: 10.1158/0008-5472.CAN-03-2770. Cancer Res. 2004. PMID: 15150106 - Overexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1.
Zhang YK, Wang H, Leng Y, Li ZL, Yang YF, Xiao FJ, Li QF, Chen XQ, Wang LS. Zhang YK, et al. Biochem Biophys Res Commun. 2011 Oct 14;414(1):233-9. doi: 10.1016/j.bbrc.2011.09.063. Epub 2011 Sep 17. Biochem Biophys Res Commun. 2011. PMID: 21951844 - Mcl-1 Is a Novel Target of miR-26b That Is Associated with the Apoptosis Induced by TRAIL in HCC Cells.
Jiang C, Long J, Liu B, Xie X, Kuang M. Jiang C, et al. Biomed Res Int. 2015;2015:572738. doi: 10.1155/2015/572738. Epub 2015 May 21. Biomed Res Int. 2015. PMID: 26078955 Free PMC article. - Mcl-1: a gateway to TRAIL sensitization.
Kim SH, Ricci MS, El-Deiry WS. Kim SH, et al. Cancer Res. 2008 Apr 1;68(7):2062-4. doi: 10.1158/0008-5472.CAN-07-6278. Cancer Res. 2008. PMID: 18381408 Review.
Cited by
- Molecular Targets in Biliary Carcinogenesis and Implications for Therapy.
Oyasiji T, Zhang J, Kuvshinoff B, Iyer R, Hochwald SN. Oyasiji T, et al. Oncologist. 2015 Jul;20(7):742-51. doi: 10.1634/theoncologist.2014-0442. Epub 2015 May 29. Oncologist. 2015. PMID: 26025932 Free PMC article. Review. - MCL-1, BCL-XL and MITF Are Diversely Employed in Adaptive Response of Melanoma Cells to Changes in Microenvironment.
Hartman ML, Talar B, Gajos-Michniewicz A, Czyz M. Hartman ML, et al. PLoS One. 2015 Jun 2;10(6):e0128796. doi: 10.1371/journal.pone.0128796. eCollection 2015. PLoS One. 2015. PMID: 26035829 Free PMC article. - Exploration of differentially expressed mRNAs and miRNAs for pediatric acute myeloid leukemia.
Wang Q, Yue C, Liu Q, Che X. Wang Q, et al. Front Genet. 2022 Sep 6;13:865111. doi: 10.3389/fgene.2022.865111. eCollection 2022. Front Genet. 2022. PMID: 36160019 Free PMC article. - Aptamer-hybrid nanoparticle bioconjugate efficiently delivers miRNA-29b to non-small-cell lung cancer cells and inhibits growth by downregulating essential oncoproteins.
Perepelyuk M, Maher C, Lakshmikuttyamma A, Shoyele SA. Perepelyuk M, et al. Int J Nanomedicine. 2016 Jul 28;11:3533-44. doi: 10.2147/IJN.S110488. eCollection 2016. Int J Nanomedicine. 2016. PMID: 27555773 Free PMC article. - MicroRNAs in Hyperglycemia Induced Endothelial Cell Dysfunction.
Silambarasan M, Tan JR, Karolina DS, Armugam A, Kaur C, Jeyaseelan K. Silambarasan M, et al. Int J Mol Sci. 2016 Apr 7;17(4):518. doi: 10.3390/ijms17040518. Int J Mol Sci. 2016. PMID: 27070575 Free PMC article.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. - PubMed
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. - PubMed
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases