TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex - PubMed (original) (raw)
TGFbeta3 inhibits E-cadherin gene expression in palate medial-edge epithelial cells through a Smad2-Smad4-LEF1 transcription complex
Ali Nawshad et al. J Cell Sci. 2007.
Abstract
Dissociation of medial-edge epithelium (MEE) during palate development is essential for mediating correct craniofacial morphogenesis. This phenomenon is initiated by TGFbeta3 upon adherence of opposing palatal shelves, because loss of E-cadherin causes the MEE seam to break into small epithelial islands. To investigate the molecular mechanisms that cause this E-cadherin loss, we isolated and cultured murine embryonic primary MEE cells from adhered or non-adhered palates. Here, we provide the first evidence that lymphoid enhancer factor 1 (LEF1), when functionally activated by phosphorylated Smad2 (Smad2-P) and Smad4 (rather than beta-catenin), binds with the promoter of the E-cadherin gene to repress its transcription in response to TGFbeta3 signaling. Furthermore, we found that TGFbeta3 signaling stimulates epithelial-mesenchymal transformation (EMT) and cell migration in these cells. LEF1 and Smad4 were found to be necessary for up-regulation of the mesenchymal markers vimentin and fibronectin, independently of beta-catenin. We proved that TGFbeta3 signaling induces EMT in MEE cells by forming activated transcription complexes of Smad2-P, Smad4 and LEF1 that directly inhibit E-cadherin gene expression.
Figures
Fig. 1
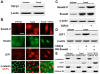
EMT is associated with nuclear localization of Smad2-P, Smad4 and LEF1, but not β-catenin. (A) Immunoblotting demonstrated that palatal adherence increases TGFβ3 expression. Unfused cells do not express TGFβ3, whereas fused cells show moderate protein expression. (B) Immunocytochemistry showed nuclear localization of Smad2-P, Smad4, and LEF1 in fused MEE cells and in fused cells treated with exogenous TGFβ3. Acquisition of the mesenchymal phenotype was observed when cells were treated with TGFβ3. β-catenin remained in the cytoplasm during EMT. Bar, 10 μm. (C) Smad2-P and total Smad2 expression was observed by immunoblotting. No expression was detected in unfused MEE cells for Smad2-P, whereas moderate levels were found in fused cells. Protein levels were heavily increased upon treatment with TGFβ3. Increased levels of total Smad2 were observed upon palatal adherence. (D) LEF1 protein expression was not detected in unfused cells, but showed steady increases in fused cells and in fused cells treated with TGFβ3. Addition of dominant-negative Smad4 prevented these increases. (E) Immunoblotting from cytoplasmic and/or membrane (Cyt/Memb) and nuclear (Nucl) protein fractions showed no evidence of nuclear β-catenin under any condition. LEF1 was observed in the nuclear fraction of fused cells, with increased levels in cells treated with TGFβ3. GAPDH and histone H3 were used as cytoplasmic and nuclear controls respectively.
Fig. 2
Confirmation of Smad and LEF1 transcriptional activities. (A) p3TP-Lux reporter gene assay demonstrated that Smad transcriptional activity increased from unfused to fused MEE cells, with a sharp increase in activity upon stimulation with TGFβ3. A dominant-negative Smad4 (DN Smad4) construct inhibited luciferase activity (mean ± s.d.; _n_=3; *P<0.05 compared with Unfused; **P<0.05 compared with p3TP-Lux). (B) pTOPFLASH-Lux (TF) reporter gene assay showed that LEF1 transcriptional activity continually increases as TGFβ3 signaling progresses. Mutated LEF1 binding sites of the pFOPFLASH-Lux reporter, as well as treatment with DN Smad4 (Smad4 being necessary for LEF1 gene expression) or dominant-negative LEF1 (DN LEF1) inhibited transcriptional activity whereas antisense β-catenin/γ-catenin oligodeoxynucleotide (AS B-catenin ODN) did not (mean ± s.d.; _n_=3; *P<0.001 compared with Unfused; **P<0.01 compared with TF).
Fig. 3
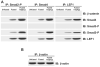
TGFβ3 signaling promotes formation of a Smad2-_P_-Smad4-LEF1 transcription complex. (A) Co-immunoprecipitation (IP) of cell extracts from MEE cells was performed using antibodies against Smad2-P, Smad4 or LEF1. Immunoblot (IB) analysis showed that Smad4, Smad2-P and LEF1 are bound together in the extracts of fused MEE cells; increased levels were observed when cells were exposed to TGFβ3. Smad3-P and β-catenin were not found in this complex. Unfused MEE cells showed no traces of interaction because LEF1 is not expressed in these cells. (B) Immunoprecipitation and immunoblotting for β-actin was used as an internal control.
Fig. 4
Smad2-_P_-Smad4-LEF1 directly bind to the promoter of the E-cadherin gene. (A) Promoter analysis of the murine E-cadherin gene revealed one unique LEF1-binding site (independent of the standard TCF and/or LEF site located in the E-pal) and a Smad-binding element (SBE), both within close proximity to the E-pal LEF1-binding region. (B) Binding of the Smad2-_P_-Smad4-LEF1 protein complex to the endogenous loci was confirmed by chromatin immunoprecipitation. Chromatin was immunoprecipitated with antibodies against IgG (negative control), LEF1, Smad4, Smad2-P or β-catenin. PCR analysis using precipitated DNA showed binding of LEF1, Smad4 and Smad2-P to LEF1 (non-E-pal), LEF1 (E-pal) and SBE regions of the E-cadherin promoter. Whereas LEF1 and Smad4 interact with their respective binding regions, all three proteins are observed because they form a transcription complex. No signal was observed from unfused MEE cells whereas a moderate signal was detected from fused MEE cells that was heavily increased upon treatment with exogenous TGFβ3. β-catenin was not detected for interaction with these binding sites.
Fig. 5
Smad2-_P_-Smad4-LEF1 directly inhibits E-cadherin gene expression. (A) Reporter gene analysis of E-cadherin promoter activity (pGL3-E-cad-Lux) demonstrated decreased expression under the influence of TGFβ3. Treatment of cells with dominant-negative Smad4 or LEF1 (DN Smad4 or DN LEF1, respectively) prevented the repression of E-cadherin, but antisense β-catenin/γ-catenin oligodeoxynucleotide (AS B-catenin ODN) did not (mean ± s.d.; _n_=3; *P<0.001 compared with Unfused; **P<0.05 compared with E-cad). (B) Site directed mutagenesis of the Smad4-binding (SBE) and LEF1-binding (E-pal and non E-pal) regions also prevented loss of E-cadherin promoter activity (mean ± s.d.; _n_=3; *P<0.001 compared with Unfused; **P<0.01 compared with E-cad). (C) Real-time quantitative PCR for E-cadherin gene expression relative to the unfused MEE negative control showed a steady decrease in fused MEE cells and fused cells treated with TGFβ3. DN-LEF1 and DN Smad4 prevented this suppression, whereas AS B-catenin ODN had no effect (mean ± s.d.; _n_=3; *P<0.001 compared with Unfused; **P<0.01 compared with + TGFβ3).
Fig. 6
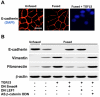
Loss of E-cadherin is associated with increased expression of mesenchymal markers. (A) Immunocytochemistry demonstrated progressive repression of E-cadherin from fused MEE cells to those exposed to exogenous TGFβ3. Bar, 10 μm. (B) E-cadherin expression was also assessed by immunoblotting, showing a moderate loss in fused cells, followed by a complete loss in fused cells treated with TGFβ3. Dominant-negative Smad4 or LEF1 (DN Smad4 or DN LEF1, respectively) prevented this repression, whereas antisense β-catenin/γ-catenin oligodeoxynucleotide (AS β-catenin ODN) had no effect. Contrarily, we found that expression of the mesenchymal markers vimentin and Fibronectin was steadily increased; DN Smad4 and DN LEF1 prevented these increases. AS β-catenin ODN had no effect on these proteins.
Fig. 7
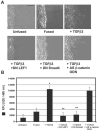
TGFβ3 signaling promotes post-EMT cell migration of MEE cells. (A) Scratch wound assays demonstrated lack of cell migration in unfused or fused MEE cells. Addition of exogenous TGFβ3 stimulated cell migration of fused cells, whereas addition of dominant-negative LEF1or Smad4 (DN LEF-1 or DN Smad4, respectively) prevented migration under these conditions. Antisense β-catenin/γ-catenin oligodeoxynucleotide (AS β-catenin ODN) did not prevent TGFβ3-induced migration. Bar, 60 μm. (B) Transwell cell migration assays showed little migration of unfused or fused MEE cells, whereas fused cells exposed to TGFβ3 were highly migratory. DN Smad4 and DN LEF1 prevented migration, but AS β-catenin ODN did not (mean ± s.d.; _n_=3; *P<0.001 compared with Unfused; **P<0.05 compared with + TGFβ3).
Fig. 8
Diagram of the proposed interaction between the Smad2-_P_-Smad4-LEF1 transcription complex and the E-cadherin gene promoter is shown to demonstrate our hypothesis of transcriptional repression.
Similar articles
- Induction of palate epithelial mesenchymal transition by transforming growth factor β3 signaling.
Jalali A, Zhu X, Liu C, Nawshad A. Jalali A, et al. Dev Growth Differ. 2012 Aug;54(6):633-48. doi: 10.1111/j.1440-169X.2012.01364.x. Epub 2012 Jul 8. Dev Growth Differ. 2012. PMID: 22775504 Free PMC article. - TGFbeta3 signaling activates transcription of the LEF1 gene to induce epithelial mesenchymal transformation during mouse palate development.
Nawshad A, Hay ED. Nawshad A, et al. J Cell Biol. 2003 Dec 22;163(6):1291-301. doi: 10.1083/jcb.200306024. J Cell Biol. 2003. PMID: 14691138 Free PMC article. - Mechanisms of transforming growth factor β induced cell cycle arrest in palate development.
Iordanskaia T, Nawshad A. Iordanskaia T, et al. J Cell Physiol. 2011 May;226(5):1415-24. doi: 10.1002/jcp.22477. J Cell Physiol. 2011. PMID: 20945347 Free PMC article. - Transforming growth factor beta (TGFbeta) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT).
Nawshad A, LaGamba D, Hay ED. Nawshad A, et al. Arch Oral Biol. 2004 Sep;49(9):675-89. doi: 10.1016/j.archoralbio.2004.05.007. Arch Oral Biol. 2004. PMID: 15275855 Review. - Regulation of epithelial-mesenchymal transition in palatal fusion.
Yu W, Ruest LB, Svoboda KK. Yu W, et al. Exp Biol Med (Maywood). 2009 May;234(5):483-91. doi: 10.3181/0812-MR-365. Epub 2009 Feb 20. Exp Biol Med (Maywood). 2009. PMID: 19234053 Review.
Cited by
- Transcriptional regulation of cell adhesion at the blood-testis barrier and spermatogenesis in the testis.
Lui WY, Cheng CY. Lui WY, et al. Adv Exp Med Biol. 2012;763:281-94. doi: 10.1007/978-1-4614-4711-5_14. Adv Exp Med Biol. 2012. PMID: 23397630 Free PMC article. Review. - Interactions between β-catenin and transforming growth factor-β signaling pathways mediate epithelial-mesenchymal transition and are dependent on the transcriptional co-activator cAMP-response element-binding protein (CREB)-binding protein (CBP).
Zhou B, Liu Y, Kahn M, Ann DK, Han A, Wang H, Nguyen C, Flodby P, Zhong Q, Krishnaveni MS, Liebler JM, Minoo P, Crandall ED, Borok Z. Zhou B, et al. J Biol Chem. 2012 Mar 2;287(10):7026-38. doi: 10.1074/jbc.M111.276311. Epub 2012 Jan 12. J Biol Chem. 2012. PMID: 22241478 Free PMC article. - Signaling pathways promoting epithelial mesenchymal transition in oral submucous fibrosis and oral squamous cell carcinoma.
Shetty SS, Sharma M, Fonseca FP, Jayaram P, Tanwar AS, Kabekkodu SP, Kapaettu S, Radhakrishnan R. Shetty SS, et al. Jpn Dent Sci Rev. 2020 Nov;56(1):97-108. doi: 10.1016/j.jdsr.2020.07.002. Epub 2020 Aug 22. Jpn Dent Sci Rev. 2020. PMID: 32874377 Free PMC article. Review. - Snail and Slug promote epithelial-mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3.
Medici D, Hay ED, Olsen BR. Medici D, et al. Mol Biol Cell. 2008 Nov;19(11):4875-87. doi: 10.1091/mbc.e08-05-0506. Epub 2008 Sep 17. Mol Biol Cell. 2008. PMID: 18799618 Free PMC article. - Tissue remodelling in chronic bronchial diseases: from the epithelial to mesenchymal phenotype.
Pain M, Bermudez O, Lacoste P, Royer PJ, Botturi K, Tissot A, Brouard S, Eickelberg O, Magnan A. Pain M, et al. Eur Respir Rev. 2014 Mar 1;23(131):118-30. doi: 10.1183/09059180.00004413. Eur Respir Rev. 2014. PMID: 24591669 Free PMC article. Review.
References
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. - PubMed
- Birchmeier W, Behrens J, Weidner KM, Hulsken J, Birchmeier C. Epithelial differentiation and the control of metastasis in carcinomas. Curr. Top. Microbiol. Immunol. 1996;213:117–135. - PubMed
- Brunet CL, Sharpe PM, Ferguson MW. Inhibition of TGF-beta 3 (but not TGF-beta 1 or TGF-beta 2) activity prevents normal mouse embryonic palate fusion. Int. J. Dev. Biol. 1995;39:345–355. - PubMed
- Carette MJ, Ferguson MW. The fate of medial edge epithelial cells during palatal fusion in vitro: an analysis by DiI labelling and confocal microscopy. Development. 1992;114:379–388. - PubMed
- Chen D, Xu W, Bales E, Colmenares C, Conacci-Sorrell M, Ishii S, Stavnezer E, Campisi J, Fisher DE, Ben-Ze′ev A, et al. SKI activates Wnt/beta-catenin signaling in human melanoma. Cancer Res. 2003;63:6626–6634. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous