Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores - PubMed (original) (raw)
Comparative Study
. 2007 May 15;21(10):1163-8.
doi: 10.1101/gad.431507.
Affiliations
- PMID: 17504936
- PMCID: PMC1865488
- DOI: 10.1101/gad.431507
Comparative Study
Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores
Emma M J King et al. Genes Dev. 2007.
Abstract
The spindle checkpoint delays anaphase onset until all chromosomes are correctly attached to microtubules. Ipl1 protein kinase (Aurora B) is required to correct inappropriate kinetochore-microtubule attachments and for the response to lack of tension between sister kinetochores. Here we identify residues in the checkpoint protein Mad3p that are phosphorylated by Ipl1p. When phosphorylation of Mad3p at two sites is prevented, the cell's response to reduced kinetochore tension is dramatically curtailed. Our data provide strong evidence for a distinct checkpoint pathway responding to lack of sister kinetochore tension, in which Ipl1p-dependent phosphorylation of Mad3p is a key step.
Figures
Figure 1.
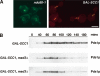
Depletion of Scc1p does not generate unattached kinetochores but promotes _MAD3_-dependent checkpoint activation. (A) Cultures of KH409 (GAL-SCC1) and SBY4341 (ndc80-1) were presynchronized in G1, and then depleted of Scc1p (KH409) or shifted to their restrictive temperature of 36°C (SBY4341). After release from G1, samples were taken at 15-min intervals for analysis of spindle (Tub1-CFP; red) and kinetochore (Mtw1-3GFP; green) positions in metaphase cells, which peaked 60 min following release. The images show cells from the 60-min time point and are maximum intensity projections of a stack of six Z-sections taken 0.2 μm apart. (B) Cultures of VBI545 (GAL-SCC1 MAD3), EK26 (GAL-SCC1 mad3Δ), and VBI560 (GAL-SCC1 mad2Δ) expressing Pds1-_myc_18 were grown in the presence of galactose, synchronized with α-factor, and then released into glucose-containing medium to allow DNA replication in the absence of SCC1 expression. Lysates were prepared from samples taken at the times indicated and immunoblotted with anti-c-myc antibody.
Figure 2.
Identification of Ipl1p-dependent phosphorylation sites in Mad3p. (A) Recombinant Mad3p was incubated with either Ipl1p or Ipl1p-D227A (kinase dead) in the presence or absence of Sli15p, together with Mg2+-[γ-32P]ATP. Phosphorylation of Mad3p was visualized following polyacrylamide gel electrophoresis and autoradiography, locating the position of the Mad3p band by Coomassie staining. (B) Following phosphorylation with Ipl1p–Sli15p complex, 32P-labeled Mad3p was digested with trypsin and separated on a C18 column, monitoring the 32P elution profile. Peaks containing 32P-labeled peptides are labeled 1–5. Peak 2 contained 57% of the applied 32P radioactivity. (C) Peak 2 was analyzed by MALDI-TOF-TOF mass spectrometry and Edman degradation: The deduced amino acid sequence from the mass spectrometric analysis is plotted against the radioactivity released in each cycle of Edman degradation. Maximal release of 32P occurred in the cycle corresponding to Ser 337. (D) _Escherichia coli_-expressed Mad3p and Mad3p-S337A were incubated with Ipl1p–Sli15p complex in the presence of Mg2+-[γ-32P]ATP. Phosphorylation of Mad3p and Mad3p-S337A was determined following electrophoresis on a polyacrylamide gel and autoradiography, locating the position of the Mad3p band by Coomassie staining. The relevant section of the stained gel is shown to confirm equal loadings. (E) The Ipl1p consensus phosphorylation site (Cheeseman et al. 2002) is shown together with the four Ipl1p phosphorylation sites identified by LC-MS with precursor ion scanning (one of which had been previously identified in C), highlighting the phosphorylated serine, the basic residue at −2, and the hydrophobic residue at +1 present at two of the sites. A phosphopeptide containing a fifth site that could not be unambiguously placed within the sequence is also shown (possible sites of phosphorylation are indicated in gray). The location of each defined phosphoserine within the Mad3p sequence is also indicated in relation to two conserved domains (Hardwick et al. 2000) involved in binding Cdc20p/Mad2p (I) and Bub3p (II).
Figure 3.
Blocking phosphorylation of Mad3p abrogates the checkpoint arrest in response to lack of tension. Cultures of EK29 (GAL-SCC1 MAD3), EK26 (GAL-SCC1 mad3Δ), EK41 [_GAL-SCC1 MAD3_-2SA (S303A, S337A)], and EK43 [_GAL-SCC1 MAD3_-4SA (S10A, S303A, S337A, S486A)] expressing Pds1-_myc_18 were grown in the presence of galactose, synchronized with α-factor, and then released into glucose-containing medium to allow DNA replication in the absence of SCC1 expression. Lysates were prepared from samples taken at the times indicated and immunoblotted with anti-c-myc antibody to detect Pds1p.
Figure 4.
Blocking phosphorylation of Mad3p does not abrogate the checkpoint arrest in response to unattached kinetochores. (A) KH173 (mad3Δ) transformed with pKH535 (MAD3), pEK089 (mad3-S303A), pEK090 (mad3-S337A), and pEK091 (mad3-2SA), was suitably diluted and spotted onto YPD agar containing the indicated benomyl concentrations. Growth is shown after 3 d at 23°C. (B) Cultures of EK45 (_PDS1-myc_18 mad3Δ) transformed with YCplac22 (empty vector), pKH535 (MAD3), or pEK091 (mad3-2SA) were synchronized with α-factor, then released into 15 μg/mL nocodazole at 23°C to activate a checkpoint arrest in response to microtubule depolymerization. Lysates were prepared from samples taken at the times indicated and were immunoblotted with anti-c-myc antibody.
Figure 5.
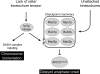
Model showing the proposed dual role of Ipl1p in promoting chromosome biorientation and checkpoint activation in response to mono-oriented chromosomes. The activation of the checkpoint machinery by lack of tension is shown as a direct response mediated through Mad3p phosphorylation by Ipl1p that is separate from the pathway that activates the checkpoint in response to unattached kinetochores, as supported by the properties of the Mad3p phosphorylation site mutants described in this study.
Similar articles
- Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores.
Keating P, Rachidi N, Tanaka TU, Stark MJ. Keating P, et al. J Cell Sci. 2009 Dec 1;122(Pt 23):4375-82. doi: 10.1242/jcs.055566. J Cell Sci. 2009. PMID: 19923271 Free PMC article. - A dual role for Bub1 in the spindle checkpoint and chromosome congression.
Meraldi P, Sorger PK. Meraldi P, et al. EMBO J. 2005 Apr 20;24(8):1621-33. doi: 10.1038/sj.emboj.7600641. Epub 2005 Mar 31. EMBO J. 2005. PMID: 15933723 Free PMC article. - Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint.
Burton JL, Solomon MJ. Burton JL, et al. Genes Dev. 2007 Mar 15;21(6):655-67. doi: 10.1101/gad.1511107. Genes Dev. 2007. PMID: 17369399 Free PMC article. - Regulation of kinetochore-microtubule attachments by Aurora B kinase.
Liu D, Lampson MA. Liu D, et al. Biochem Soc Trans. 2009 Oct;37(Pt 5):976-80. doi: 10.1042/BST0370976. Biochem Soc Trans. 2009. PMID: 19754435 Review. - Attachment and tension in the spindle assembly checkpoint.
Zhou J, Yao J, Joshi HC. Zhou J, et al. J Cell Sci. 2002 Sep 15;115(Pt 18):3547-55. doi: 10.1242/jcs.00029. J Cell Sci. 2002. PMID: 12186941 Review.
Cited by
- Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation.
Wang F, Ulyanova NP, Daum JR, Patnaik D, Kateneva AV, Gorbsky GJ, Higgins JM. Wang F, et al. J Cell Biol. 2012 Oct 15;199(2):251-68. doi: 10.1083/jcb.201205106. J Cell Biol. 2012. PMID: 23071152 Free PMC article. - Identification of a Sgo2-Dependent but Mad2-Independent Pathway Controlling Anaphase Onset in Fission Yeast.
Meadows JC, Lancaster TC, Buttrick GJ, Sochaj AM, Messin LJ, Del Mar Mora-Santos M, Hardwick KG, Millar JBA. Meadows JC, et al. Cell Rep. 2017 Feb 7;18(6):1422-1433. doi: 10.1016/j.celrep.2017.01.032. Cell Rep. 2017. PMID: 28178520 Free PMC article. - Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore.
Foley EA, Kapoor TM. Foley EA, et al. Nat Rev Mol Cell Biol. 2013 Jan;14(1):25-37. doi: 10.1038/nrm3494. Nat Rev Mol Cell Biol. 2013. PMID: 23258294 Free PMC article. Review. - Overcoming inhibition in the spindle checkpoint.
Vanoosthuyse V, Hardwick KG. Vanoosthuyse V, et al. Genes Dev. 2009 Dec 15;23(24):2799-805. doi: 10.1101/gad.1882109. Genes Dev. 2009. PMID: 20008930 Free PMC article. Review. - A brief history of error.
Murray AW. Murray AW. Nat Cell Biol. 2011 Oct 3;13(10):1178-82. doi: 10.1038/ncb2348. Nat Cell Biol. 2011. PMID: 21968991
References
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G., Chan C.S., Drubin D.G., Barnes G., Drubin D.G., Barnes G., Barnes G. Phospho-regulation of kinetochore–microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. - PubMed
- Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K., Michaelis C., Shevchenko A., Mann M., Nasmyth K., Shevchenko A., Mann M., Nasmyth K., Mann M., Nasmyth K., Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. - PubMed
- Dewar H., Tanaka K., Nasmyth K., Tanaka T.U., Tanaka K., Nasmyth K., Tanaka T.U., Nasmyth K., Tanaka T.U., Tanaka T.U. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. - PubMed
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S., Johnson T., Mortlock A., Keen N., Taylor S.S., Mortlock A., Keen N., Taylor S.S., Keen N., Taylor S.S., Taylor S.S. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and CENP-E to kinetochores. J. Cell Biol. 2003;161:267–280. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases