A MicroRNA feedback circuit in midbrain dopamine neurons - PubMed (original) (raw)
A MicroRNA feedback circuit in midbrain dopamine neurons
Jongpil Kim et al. Science. 2007.
Abstract
MicroRNAs (miRNAs) are evolutionarily conserved, 18- to 25-nucleotide, non-protein coding transcripts that posttranscriptionally regulate gene expression during development. miRNAs also occur in postmitotic cells, such as neurons in the mammalian central nervous system, but their function is less well characterized. We investigated the role of miRNAs in mammalian midbrain dopaminergic neurons (DNs). We identified a miRNA, miR-133b, that is specifically expressed in midbrain DNs and is deficient in midbrain tissue from patients with Parkinson's disease. miR-133b regulates the maturation and function of midbrain DNs within a negative feedback circuit that includes the paired-like homeodomain transcription factor Pitx3. We propose a role for this feedback circuit in the fine-tuning of dopaminergic behaviors such as locomotion.
Figures
Figure 1
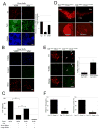
Dicer is essential for terminal differentiation of ES cells to a DN phenotype. (A) Floxed Dicer conditional knockout cells were transduced with CRE lentivirus at EB stage 4 and analyzed by immunostaining at day 7 of stage 5 with antibodies specific for TH (Red), TujI (Green) and GABA (blue). Cultures transduced with a lentiviral Cre vector (vCre) are essentially devoid of TH+ neurons, whereas TujI+ and GABA+ cells are reduced by approximately 40%-60%. (Scale bars, 100μm). (n=3 independent samples per group; 2 additional independent sets of samples showed consistent results). Data represent means ± S.E.M. ANOVA-test, *p < 0.05. (C) The Dicer deletion phenotype, as in (A) can be ‘rescued’ by transfection of midbrain-derived small RNAs (<200bp) not large RNAs (>200bp) (Two independent experiments of 3 sets each with 10 visual fields per set), Data represent means ± S.E.M. ANOVA-test, **p < 0.01. (D) Immunostaining of brain sections from 8 week old DATCRE/+ :Dicerflox/flox mice for tyrosine hydroxylase (TH) demonstrates loss of approximately 90% of midbrain DNs in the substantia nigra (SN) and ventral tegmental area (VTA) and their axonal projections to the striatum relative to control littermates (DATCRE/+ : Dicerflox/+; N=3 for each genotype). (Scale bars,200μm) (F) Locomotor activity of DATCRE/+ :Dicerflox/flox mice in the open field. The total distance traveled during the test was significantly decreased in DATCRE/+ :Dicerflox/flox mice. (N=4 for each genotype), Data represent means ± S.E.M. Student's t test, *p < 0.05, **p < 0.01.
Figure 2
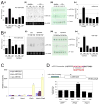
miR-133b is enriched in midbrain and is deficient in PD samples. (A) Expression analysis for precursor miR-133b in cerebral cortex (CX), midbrain (MB), or cerebellum (CB) of healthy human and Parkinson's disease (PD) patient brain. RNA protection assay for miR133a1 and miR133b expression from normal or PD human brain and quantification of RNA protection assay. n=3 independent experiments; data represent means ± S.E.M. ANOVA test, *p < 0.05. (B) Expression analysis for precursor miR-133b in cerebral cortex (CX), midbrain (MB), or cerebellum (CB) of control mice and Aphakia mutant. RNA protection assay for miR133a1 and miR133b expression from control mice and Aphakia mutant mice brain and quantification of RNA protection assay. n=3 independent experiments; data represent means ± S.E.M. ANOVA test, *p < 0.05. (C) qPCR analysis of murine ES cultures differentiated by EB and transduced at stage 4 with viral vectors for Nurr1, Pitx3, both, or GFP vector control. Pitx3 transduction (but not Nurr1 transduction) leads to the specific induction of miR-133b precursor expression assayed at stage 5; miR-133a1 and miR-133a2 precursors are not induced by Pitx3 overexpression. n=3 independent experiments; data represent means ± S.E.M. ANOVA test, *p < 0.05.
Figure 3
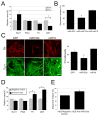
miR-133b suppresses dopamine neuron terminal differentiation and function. (A) Overexpression of miR-133b precursor in primary embryonic rat midbrain cultures leads to decreased expression of dopamine neuron mRNAs including TH and DAT, whereas Nurr1 and Pitx3 mRNAs are not altered. Data represent means ± S.E.M. n=3 independent experiments; Student's t test, *p < 0.05. (B) Depolarization-induced dopamine release was quantified in cultures transduced with miR-133b precursor vector or GFP control. miR-133b overexpression reduced dopamine release in murine ES cultures derived dopamine neurons. Data represent means ± S.E.M. n=5; ANOVA test, *p < 0.05. (C) Overexpression of miR-133b during EB stage 4 of mouse ES cell differentiation resulted in a significant decrease in TH-positive cells. Data represent means ± S.E.M. n=3 independent experiments; ANOVA test, *p < 0.05. (D) Knockdown of miR-133b by penetratin conjugated antisense miR133b 2OM oligo in primary embryonic rat midbrain cultures leads to increased expression of dopamine neuron mRNAs including TH and DAT, whereas Nurr1 and Pitx3 mRNAs are not significantly altered. Data represent means ± S.E.M. n=5 independent experiments; Student's t test, *p < 0.05. (E) Depolarization-induced dopamine release was quantified in cultures with miR-133b knockdown or oligonucleotide control. miR-133b knockdown induced dopamine release in murine ES cultures. Data represent means ± S.E.M. n=3 independent experiments; Student's t test, *p < 0.05.
Figure 4
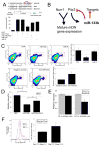
Pitx3 is a target of miR-133b activity. (A) Upper Panel: Schematic of a luciferase assay vector, pGL3-Pitx3 and pGL3-Mut-Pitx3 that harbors 300bp Pitx3 3′-UTR sequences predicted to be subject to miR-133b regulation. Lower panel: Overexpression of miR-133b (but not miR-18) in 293 cells decreases luciferase expression from pGL3-Pitx3 (but not control vector and pGL3-mut Pitx3). Data represent means ± S.E.M. n=3 independent experiments; Student's t test, *p < 0.05. (B) miR-133b and Pitx3 define a negative feedback loop in midbrain DN function and differentiation. It is likely that miR-133b has additional significant targets. (C) Upper panels: FACS analysis of primary midbrain cultures transduced with miR-133b or control vectors using antibodies for Pitx3 and TH reveals that miR-133b overexpression decrease in both Pitx3 and TH protein expression. Pitx3 is reduced in TH-positive cells transduced with miR-133b relative to control vector or miR-18. Data represent means ± S.E.M. n=3 independent experiments; ANOVA test, *p < 0.05. Lower panels: Knockdown of miR-133b using modified oligonucleotides leads to a significant induction in Pitx3 and TH protein levels relative to control oligonucleotides. Data represent means ± S.E.M. n=3 independent experiments; Student's t test, *p < 0.05. (E) miR-133b inhibition by modified oligonucleotides in Pitx3-deficient Aphakia primary neuron cultures fails to induce TH or DAT transcription. Data represent means ± S.E.M. n=3 independent experiments; Student's t test, *p < 0.05.
Comment in
- Molecular biology. miRNAs in neurodegeneration.
Hébert SS, De Strooper B. Hébert SS, et al. Science. 2007 Aug 31;317(5842):1179-80. doi: 10.1126/science.1148530. Science. 2007. PMID: 17761871 No abstract available.
Similar articles
- An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool.
Anderegg A, Lin HP, Chen JA, Caronia-Brown G, Cherepanova N, Yun B, Joksimovic M, Rock J, Harfe BD, Johnson R, Awatramani R. Anderegg A, et al. PLoS Genet. 2013;9(12):e1003973. doi: 10.1371/journal.pgen.1003973. Epub 2013 Dec 12. PLoS Genet. 2013. PMID: 24348261 Free PMC article. - Normal midbrain dopaminergic neuron development and function in miR-133b mutant mice.
Heyer MP, Pani AK, Smeyne RJ, Kenny PJ, Feng G. Heyer MP, et al. J Neurosci. 2012 Aug 8;32(32):10887-94. doi: 10.1523/JNEUROSCI.1732-12.2012. J Neurosci. 2012. PMID: 22875923 Free PMC article. - Molecular biology. miRNAs in neurodegeneration.
Hébert SS, De Strooper B. Hébert SS, et al. Science. 2007 Aug 31;317(5842):1179-80. doi: 10.1126/science.1148530. Science. 2007. PMID: 17761871 No abstract available. - The role of transcription factor Pitx3 in dopamine neuron development and Parkinson's disease.
Li J, Dani JA, Le W. Li J, et al. Curr Top Med Chem. 2009;9(10):855-9. Curr Top Med Chem. 2009. PMID: 19754401 Free PMC article. Review. - The Crucial Roles of Pitx3 in Midbrain Dopaminergic Neuron Development and Parkinson's Disease-Associated Neurodegeneration.
Wang X, Chen X, Liu G, Cai H, Le W. Wang X, et al. Int J Mol Sci. 2023 May 11;24(10):8614. doi: 10.3390/ijms24108614. Int J Mol Sci. 2023. PMID: 37239960 Free PMC article. Review.
Cited by
- MicroRNAs: Key Regulators in the Central Nervous System and Their Implication in Neurological Diseases.
Cao DD, Li L, Chan WY. Cao DD, et al. Int J Mol Sci. 2016 May 28;17(6):842. doi: 10.3390/ijms17060842. Int J Mol Sci. 2016. PMID: 27240359 Free PMC article. Review. - Overexpression of MicroRNA-133a Inhibits Apoptosis and Autophagy in a Cell Model of Parkinson's Disease by Downregulating Ras-Related C3 Botulinum Toxin Substrate 1 (RAC1).
Lu W, Lin J, Zheng D, Hong C, Ke L, Wu X, Chen P. Lu W, et al. Med Sci Monit. 2020 Jul 27;26:e922032. doi: 10.12659/MSM.922032. Med Sci Monit. 2020. PMID: 32713934 Free PMC article. - MicroRNA 433 regulates nonsense-mediated mRNA decay by targeting SMG5 mRNA.
Jin Y, Zhang F, Ma Z, Ren Z. Jin Y, et al. BMC Mol Biol. 2016 Jul 29;17(1):17. doi: 10.1186/s12867-016-0070-z. BMC Mol Biol. 2016. PMID: 27473591 Free PMC article. - MicroRNAs: regulators of neuronal fate.
Sun AX, Crabtree GR, Yoo AS. Sun AX, et al. Curr Opin Cell Biol. 2013 Apr;25(2):215-21. doi: 10.1016/j.ceb.2012.12.007. Epub 2013 Jan 29. Curr Opin Cell Biol. 2013. PMID: 23374323 Free PMC article. Review. - Alzheimer's disease models and functional genomics-How many needles are there in the haystack?
Götz J, Matamales M, Götz NN, Ittner LM, Eckert A. Götz J, et al. Front Physiol. 2012 Aug 8;3:320. doi: 10.3389/fphys.2012.00320. eCollection 2012. Front Physiol. 2012. PMID: 22934069 Free PMC article.
References
- He L, Hannon GJ. Nat Rev Genet. 2004 Jul;5:522. - PubMed
- Meister G, Tuschl T. Nature. 2004 Sep 16;431:343. - PubMed
- Ambros V. Nature. 2004 Sep 16;431:350. - PubMed
- Bartel DP. Cell. 2004 Jan 23;116:281. - PubMed
- Lee Y, et al. Nature. 2003 Sep 25;425:415. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases