Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks - PubMed (original) (raw)
Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks
Haico van Attikum et al. EMBO J. 2007.
Abstract
INO80 and SWR1 are two closely related ATP-dependent chromatin remodeling complexes that share several subunits. Ino80 was reported to be recruited to the HO endonuclease-induced double-strand break (DSB) at the budding yeast mating-type locus, MAT. We find Swr1 similarly recruited in a manner dependent on the phosphorylation of H2A (gammaH2AX). This is not unique to cleavage at MAT; both Swr1 and Ino80 bind near an induced DSB on chromosome XV. Whereas Swr1 incorporates the histone variant H2A.Z into chromatin at promoters, H2A.Z levels do not increase at DSBs. Instead, H2A.Z, gammaH2AX and core histones are coordinately removed near the break in an INO80-dependent, but SWR1-independent, manner. Mutations in INO80-specific subunits Arp8 or Nhp10 impair the binding of Mre11 nuclease, yKu80 and ATR-related Mec1 kinase at the DSB, resulting in defective end-processing and checkpoint activation. In contrast, Mre11 binding, end-resection and checkpoint activation were normal in the swr1 strain, but yKu80 loading and error-free end-joining were impaired. Thus, these two related chromatin remodelers have distinct roles in DSB repair and checkpoint activation.
Figures
Figure 1
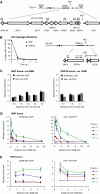
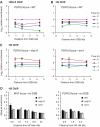
Ino80 and Swr1 are recruited to chromosomal DNA DSBs. (A, B) Schematic representations of genes and rt–PCR primer sets on chromosome III (A) and XV (B) in the haploid yeast strain JKM179 and its derivative GA3632. Potential donor loci HML and HMR are deleted. The cleavage efficiency at MAT and PDR10 upon induction of HO is shown over time. (C) Absolute fold enrichment of Ino80-Myc and Swr1-HA ChIPs at the indicated distances from the HO site at MAT and PDR10 for strains without a DSB. Cells were grown in the presence of glucose to repress HO. (D) Relative fold enrichment of Ino80-Myc and Swr1-HA ChIPs at the indicated distances from the HO cut site at MAT, and the indicated times on galactose. The value at 0 h is arbitrarily set as 1. Cleavage efficiencies were 96, 98 and 99% (Ino80-Myc ChIP), and 89, 98, 99% (Swr1-HA ChIP) at 1, 2 and 4 h after HO induction, respectively. (E) As panel D, except that this shows Ino80-Myc and Swr1-HA ChIPs at the DSB in the PDR10 locus. Cleavage efficiencies were 93, 97 and 99% (Ino80-Myc ChIP), and 89, 98, 99% (Swr1-Myc ChIP).
Figure 2
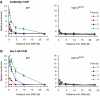
H2AX phosphorylation at sites near a DSB at MAT is required for recruitment of INO80 and SWR1. (A) Relative fold enrichment of Ino80-Myc ChIP for WT and hta1/2 S129* strains at the indicated distances from the HO cut site, and the indicated times on galactose. The enrichment is corrected for cleavage efficiencies, which were 96, 98 and 99% (WT), and 89, 98, 99% (hta1/2 S129*). (B) As panel A, except that this shows ChIP for Swr1-HA. Cleavage efficiencies were 89, 98 and 99% (WT), and 78, 92, 95% (hta1/2 S129*).
Figure 3
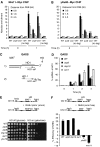
H2A.Z is evicted from sites near a DSB at MAT and PDR10. (A) Absolute fold enrichment of H2A.Z ChIP at the indicated distances from the HO site at MAT for a WT strain with and without DSB, and a swr1 mutant without DSB is shown. Cells were grown in the presence of glucose to repress HO, or in the presence of galactose for 4 h to induce HO. Cleavage efficiency was 99% at 4 h after HO induction. (B) As panel A, except that this shows H2A.Z ChIP at the DSB in the PDR10 locus for a WT strain. Cleavage efficiency was 97%. (C) As panel A, except that this shows H2A.Z ChIP at MATinc for a WT strain. Cells were grown in the presence of glucose to repress HO or in the presence of galactose for 4 h to induce HO. On galactose no DSB induction at MATinc occurs. (D) As panel C, except that this shows histone H3 ChIP.
Figure 4
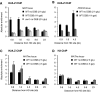
INO80 is involved in γH2AX, H2A.Z and histone H3 eviction from sites near a DSB at MAT. (A) Relative fold enrichment of γH2AX ChIP at the indicated distances from the HO cut site at MAT, and the indicated times on galactose for WT, nhp10, and swr1 strains is shown as in Figure 1. Cleavage efficiencies were 90, 98 and 99% (WT), 44, 75 and 95% (nhp10), and 76, 91 and 96% (swr1). (B) As panel A, except that this shows H2A.Z ChIP for WT, nhp10 and hta1/2 S129* strains. Cleavage efficiencies were 92, 97 and 99% (WT), 62, 83 and 94% (nhp10), and 88, 96 and 97% (hta1/2 S129*). (C) As panel A, except that this shows histone H3 ChIP for WT, nhp10 and swr1 strains. Cleavage efficiencies were 89, 96 and 98% (WT), 46, 79 and 94% (nhp10), and 89, 96 and 98% (swr1).
Figure 5
H2A.Z and histone H3 are evicted from sites near a DSB at PDR10. (A) Relative fold enrichment of H2A.Z ChIP at the indicated distances from the HO cut site at PDR10, and the indicated times on galactose for a WT strain is shown as in Figure 1. Cleavage efficiencies were 84, 95 and 97%. (B) As panel A, except that this shows histone H3 ChIP. Cleavage efficiencies were 87, 92 and 94%. (C) As panel B, except that this shows histone H3 ChIP for nhp10 and swr1 strains. Cleavage efficiencies were 72, 88 and 91% (nhp10), and 89, 96 and 98% (swr1). (D) Absolute fold enrichment of histone H3 ChIP at the indicated distances from the HO site at MAT and PDR10 for WT, nhp10 and swr1 strains in the absence of a DSB. Cells were grown in the presence of glucose to repress HO.
Figure 6
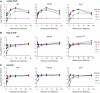
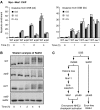
Effect of ARP8, NHP10 and SWR1 deletion on Mre11 and yKu80 recruitment, end-resection and non-homologous end-joining. (A) Absolute fold enrichment of Mre11–Myc ChIP at the indicated distances from the HO cut site at MAT at 0 and 1 h on galactose. The enrichment is corrected for cleavage efficiencies, which were 86% (WT), 84% (arp8), 86% (swr1) and 96% (WT no tag). (B) As panel A, except that this shows ChIP for yKu80–Myc. Cleavage efficiencies were 98% (WT), 85% (arp8), 90% (swr1), and 96% (WT no tag). (C) Schematic representation of the QAOS assay (see text). DNA template molecule, generated using primer HO1-ss, is quantified by rt–PCR using primers HO1-f and Tag. (D) QAOS assay for ssDNA formed at the HO1 site at the indicated times on galactose in WT, arp8, nhp10 and swr1 strains. The amount of ssDNA is corrected for cleavage efficiencies, which were 92, 97, 99% (WT), 56, 78, 85% (arp8), 48, 79, 93% (nhp10) and 77, 90, 96% (swr1). (E) Error-prone NHEJ repair of a DSB at MAT is slightly reduced in mutants in INO80, but not SWR1. Repair of the DSB at MAT can only occur by NHEJ. Error-prone NHEJ was monitored by scoring survival of the indicated strains after plating serial 10-fold dilutions on medium with either 2% glucose or galactose. (F) Error-free NHEJ repair of a DSB at MAT is reduced in swr1, but not in arp8 or nhp10 mutants. NHEJ proficiency was determined by rt–PCR, using primers that span the DSB at MAT or anneal to the SMC2 control locus. HO was repressed (_t_=0) and induced for 1 h (t_=1). After 1 h, cells expressing HO were shifted to a glucose medium and incubated for 2 h (t_=3) to allow error-free NHEJ. Error-free end-joining efficiency was calculated from PCR cycle threshold values for MAT (ct_MAT) normalized to SMC2 (ct_SMC2) from _t_=0, _t_=1 and _t_=3, using the following formula:  . Each point represents the average of two independent experiments. Standard deviation is shown.
. Each point represents the average of two independent experiments. Standard deviation is shown.
Figure 7
Deletion of ARP8, but not SWR1, reduces Mec1 recruitment and checkpoint activation in response to a DSB at MAT. (A) Absolute fold enrichment of Myc–Mec1 ChIP for WT and arp8 strains (left panel) and WT and swr1 strains (right panel) is shown as in Figure 1. Cleavage efficiencies were 89, 97 and 99% (WT, left panel), 56, 78 and 85% (arp8), 95, 98 and 99% (WT, right panel), and 85, 95 and 97% (swr1). (B) Rad53 kinase activation was determined by Western blot analysis in WT, arp8, swr1 and rad9rad24 strains. (C) Representation of INO80- and SWR1-dependent events at a DSB. INO80 removes histone H2A variants (γH2AX and H2A.Z) and core histones, facilitating Mre11 binding, end-processing, Mec1 recruitment and checkpoint activation. In contrast, SWR1 does not affect histone loss, Mre11 binding or end-processing, but is required for yKu80 binding, as is INO80. Whereas SWR1 facilitates error-free NHEJ, INO80 only plays a minor role in error-prone NHEJ.
Similar articles
- The INO80 complex is required for damage-induced recombination.
Kawashima S, Ogiwara H, Tada S, Harata M, Wintersberger U, Enomoto T, Seki M. Kawashima S, et al. Biochem Biophys Res Commun. 2007 Apr 13;355(3):835-41. doi: 10.1016/j.bbrc.2007.02.036. Epub 2007 Feb 15. Biochem Biophys Res Commun. 2007. PMID: 17320816 - Chromatin remodeling in DNA double-strand break repair.
Bao Y, Shen X. Bao Y, et al. Curr Opin Genet Dev. 2007 Apr;17(2):126-31. doi: 10.1016/j.gde.2007.02.010. Epub 2007 Feb 22. Curr Opin Genet Dev. 2007. PMID: 17320375 Review. - Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break.
Kalocsay M, Hiller NJ, Jentsch S. Kalocsay M, et al. Mol Cell. 2009 Feb 13;33(3):335-43. doi: 10.1016/j.molcel.2009.01.016. Mol Cell. 2009. PMID: 19217407 - ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex.
Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. Mizuguchi G, et al. Science. 2004 Jan 16;303(5656):343-8. doi: 10.1126/science.1090701. Epub 2003 Nov 26. Science. 2004. PMID: 14645854 - Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes.
Morrison AJ, Shen X. Morrison AJ, et al. Nat Rev Mol Cell Biol. 2009 Jun;10(6):373-84. doi: 10.1038/nrm2693. Epub 2009 May 8. Nat Rev Mol Cell Biol. 2009. PMID: 19424290 Free PMC article. Review.
Cited by
- Biochemical mechanism of DSB end resection and its regulation.
Daley JM, Niu H, Miller AS, Sung P. Daley JM, et al. DNA Repair (Amst). 2015 Aug;32:66-74. doi: 10.1016/j.dnarep.2015.04.015. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956866 Free PMC article. Review. - Chromatin modifications and the DNA damage response to ionizing radiation.
Kumar R, Horikoshi N, Singh M, Gupta A, Misra HS, Albuquerque K, Hunt CR, Pandita TK. Kumar R, et al. Front Oncol. 2013 Jan 22;2:214. doi: 10.3389/fonc.2012.00214. eCollection 2012. Front Oncol. 2013. PMID: 23346550 Free PMC article. - To trim or not to trim: progression and control of DSB end resection.
Granata M, Panigada D, Galati E, Lazzaro F, Pellicioli A, Plevani P, Muzi-Falconi M. Granata M, et al. Cell Cycle. 2013 Jun 15;12(12):1848-60. doi: 10.4161/cc.25042. Epub 2013 May 29. Cell Cycle. 2013. PMID: 23708517 Free PMC article. - The yeast high mobility group protein HMO2, a subunit of the chromatin-remodeling complex INO80, binds DNA ends.
Ray S, Grove A. Ray S, et al. Nucleic Acids Res. 2009 Oct;37(19):6389-99. doi: 10.1093/nar/gkp695. Epub 2009 Sep 2. Nucleic Acids Res. 2009. PMID: 19726587 Free PMC article. - BZLF1 interacts with chromatin remodelers promoting escape from latent infections with EBV.
Schaeffner M, Mrozek-Gorska P, Buschle A, Woellmer A, Tagawa T, Cernilogar FM, Schotta G, Krietenstein N, Lieleg C, Korber P, Hammerschmidt W. Schaeffner M, et al. Life Sci Alliance. 2019 Mar 29;2(2):e201800108. doi: 10.26508/lsa.201800108. Print 2019 Apr. Life Sci Alliance. 2019. PMID: 30926617 Free PMC article.
References
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF (2002) Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415 - PubMed
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ (2001) ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 276: 42462–42467 - PubMed
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J (2004) Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell 16: 979–990 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous