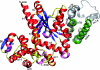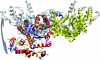Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of mimivirus TyrRS and MetRS - PubMed (original) (raw)
Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of mimivirus TyrRS and MetRS
Chantal Abergel et al. J Virol. 2007 Nov.
Abstract
Aminoacyl-tRNA synthetases are pivotal in determining how the genetic code is translated in amino acids and in providing the substrate for protein synthesis. As such, they fulfill a key role in a process universally conserved in all cellular organisms from their most complex to their most reduced parasitic forms. In contrast, even complex viruses were not found to encode much translation machinery, with the exception of isolated components such as tRNAs. In this context, the discovery of four aminoacyl-tRNA synthetases encoded in the genome of mimivirus together with a full set of translation initiation, elongation, and termination factors appeared to blur what was once a clear frontier between the cellular and viral world. Functional studies of two mimivirus tRNA synthetases confirmed the MetRS specificity for methionine and the TyrRS specificity for tyrosine and conformity with the identity rules for tRNA(Tyr) for archea/eukarya. The atomic structure of the mimivirus tyrosyl-tRNA synthetase in complex with tyrosinol exhibits the typical fold and active-site organization of archaeal-type TyrRS. However, the viral enzyme presents a unique dimeric conformation and significant differences in its anticodon binding site. The present work suggests that mimivirus aminoacyl-tRNA synthetases function as regular translation enzymes in infected amoebas. Their phylogenetic classification does not suggest that they have been acquired recently by horizontal gene transfer from a cellular host but rather militates in favor of an intricate evolutionary relationship between large DNA viruses and ancestral eukaryotes.
Figures
FIG. 1.
Functional assays of TyrRSapm and MetRSapm. Amino acid activation of TyrRSapm and MetRSapm. Reactions were conducted in the presence of a mix of all amino acids minus (□) or plus (▪) tyrosine (A) or methionine (B). (C) Tyrosylation of E. coli (▵) and yeast (▴) native tRNATyr by TyrRSapm. Enzyme and tRNA concentrations were 20 μM and 1.3 μM, respectively.
FIG. 2.
Invariance of the tyrosinol binding site. Amino acids closest to the tyrosinol molecule (A) in the TyrRSapm and (B) in the M. jannaschii TyrRS structures (1J1U).
FIG. 3.
Structure-based alignment of TyrRSs. TyrRSapm (2J5B) was aligned with eukaryal (1Q11, human, core structure) and archaeal (2CYC, P. horikoshii; 2CYA, Aeropyrum pernix; 2CYB, Archaeoglobus fulgidus; 1J1U and 1U7D, M. jannaschii complex and apo form) structures. The closest TyrRSapm homologues from protozoa and plants are also included (EHISTO, Entamoeba histolytica; DICDIS, Dictyostelium discoideum; OSATI, Oryza sativa; PYOELI, Plasmodium yoelii; PFALC, Plasmodium falciparum; ATHAL, Arabidopsis thaliana; CHOMI, Cryptosporidium hominis; GLAMB, Giardia lamblia). The secondary-structure elements of TyrRSapm and M. jannaschii are, respectively, indicated above and below the multiple alignment. The N-terminal, Rossmann fold, CP1, and C-terminal domains are colored in pink, blue, green, and red, respectively. Strictly conserved residues are boxed in red. Residues involved in tyrosine binding (Fig. 2) are highlighted in gray. This alignment was produced with 3DCoffee (
http://www.igs.cnrs-mrs.fr/Tcoffee/tcoffee\_cgi/index.cgi
) (48), and the figure was produced with ESPript (31).
FIG. 4.
Cartoon comparison of the dimer interface. The two α7 helices of TyrRSapm are in green, and the conserved αs6-turn-αs7 structural motif (found in other TyrRS dimers) is colored in silver, transparent for the first monomer and opaque for the second one. This figure illustrates the nearly 90° rotation of the TyrRSapm second monomer (colored according to secondary-structure elements: red, α-helices; blue, β-strands; yellow, coils and turns; pink, η helices). N-term, N terminus; C-term, C terminus.
FIG. 5.
Cartoon representation of the TyrRSapm dimer superimposed on the archaeal TyrRS 2CYC from P. horikoshii. The archaeal dimer is transparent and colored in silver, except for the structural motif (αs6-turn-αs7, corresponding to the α7 helix in TyrRSapm), colored in opaque cyan. The TyrRSapm first monomer superimposed on the 1CYC first monomer is colored in yellow. The second monomer secondary-structure elements are colored as outlined in the legend for Fig. 4. The arrow shows the rotation to be applied to the mimivirus second monomer to superimpose its β7-turn-β8 onto the P. horikoshii βs7-ηs3-βs8 motif. N-term, N terminus; C-term, C terminus.
FIG. 6.
Superimposition of the C-terminal anticodon binding domain of TyrRSapm (solid) on the M. jannaschii TyrRS/tRNA complex (transparent). The anticodon appears solid in the transparent tRNA surface.
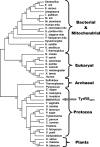
FIG. 7.
Phylogenetic position of mimivirus TyrRS. S. aureus, Staphylococcus aureus; T. pallidum, Treponema pallidum; M. loti, Mesorhizobium loti; M. penetrans, Mycoplasma penetrans; S. pombe, Schizosaccharomyces pombe; C. elegans, Caenorhabditis elegans; B. subtilis, Bacillus subtilis; A. aeoliticus, Aquifex aeoliticus; P. carinii, Pneumocystis carinii; E. cuniculi, Encephalitozoon cuniculi; D. melanogaster, Drosophila melanogaster; X. laevis, Xenopus laevis; E. dispar, Entamoeba dispar; E. invadens, Entamoeba invadens; P. ramorum, Phytophthora ramorum; P. sojae, Phytophthora sojae; C. parvum, Cryptosporidium parvum; C. hominis, Cryptosporidium hominis; P. yoelii, Plasmodium yoelii; O. sativa, Oryza sativa; A. thaliana, Arabidopsis thaliana; N. tabacum, Nicotiana tabacum. All other organisms are defined in the text. mito, mitochondrial sequences.
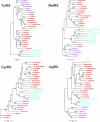
FIG. 8.
Phylogenetic position of mimivirus aaRS. Archeal sequences are colored in purple, eukaryotic in red, mitochondrial (Mito) (bacterial type) in cyan, and mimivirus in green. A. fulgidus, Archaeoglobus fulgidus; P. abyssi, Pyrococcus abyssi; A. pernix, Aeropyrum pernix; T. volcanium, Thermoplasma volcanium; A. gambiae, Anopheles gambiae; G. gallus, Gallus gallus; B. taurus, Bos taurus; H. sapiens, Homo sapiens; D. rerio, Danio rerio; X. tropicalis, Xenopus tropicalis. All other organisms are defined in the text or the legend for Fig. 7.
Similar articles
- Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion.
Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S. Kobayashi T, et al. Nat Struct Biol. 2003 Jun;10(6):425-32. doi: 10.1038/nsb934. Nat Struct Biol. 2003. PMID: 12754495 - Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae.
Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. Arslan D, et al. Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17486-91. doi: 10.1073/pnas.1110889108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987820 Free PMC article. - Studies on crenarchaeal tyrosylation accuracy with mutational analyses of tyrosyl-tRNA synthetase and tyrosine tRNA from Aeropyrum pernix.
Iwaki J, Endo K, Ichikawa T, Suzuki R, Fujimoto Z, Momma M, Kuno A, Nishimura S, Hasegawa T. Iwaki J, et al. J Biochem. 2012 Dec;152(6):539-48. doi: 10.1093/jb/mvs114. Epub 2012 Sep 29. J Biochem. 2012. PMID: 23024156 - Discrimination between transfer-RNAs by tyrosyl-tRNA synthetase.
Bedouelle H, Guez-Ivanier V, Nageotte R. Bedouelle H, et al. Biochimie. 1993;75(12):1099-108. doi: 10.1016/0300-9084(93)90009-h. Biochimie. 1993. PMID: 8199245 Review. - Evolution of the tRNA(Tyr)/TyrRS aminoacylation systems.
Bonnefond L, Giegé R, Rudinger-Thirion J. Bonnefond L, et al. Biochimie. 2005 Sep-Oct;87(9-10):873-83. doi: 10.1016/j.biochi.2005.03.008. Epub 2005 Apr 8. Biochimie. 2005. PMID: 16164994 Review.
Cited by
- Dance with the Devil: Stress Granules and Signaling in Antiviral Responses.
Eiermann N, Haneke K, Sun Z, Stoecklin G, Ruggieri A. Eiermann N, et al. Viruses. 2020 Sep 4;12(9):984. doi: 10.3390/v12090984. Viruses. 2020. PMID: 32899736 Free PMC article. Review. - Horizontal gene transfers with or without cell fusions in all categories of the living matter.
Sinkovics JG. Sinkovics JG. Adv Exp Med Biol. 2011;714:5-89. doi: 10.1007/978-94-007-0782-5_2. Adv Exp Med Biol. 2011. PMID: 21506007 Free PMC article. Review. - Two classes of EF1-family translational GTPases encoded by giant viruses.
Zinoviev A, Kuroha K, Pestova TV, Hellen CUT. Zinoviev A, et al. Nucleic Acids Res. 2019 Jun 20;47(11):5761-5776. doi: 10.1093/nar/gkz296. Nucleic Acids Res. 2019. PMID: 31216040 Free PMC article. - Giant Viruses of Amoebas: An Update.
Aherfi S, Colson P, La Scola B, Raoult D. Aherfi S, et al. Front Microbiol. 2016 Mar 22;7:349. doi: 10.3389/fmicb.2016.00349. eCollection 2016. Front Microbiol. 2016. PMID: 27047465 Free PMC article. Review. - Virologs, viral mimicry, and virocell metabolism: the expanding scale of cellular functions encoded in the complex genomes of giant viruses.
Moniruzzaman M, Erazo Garcia MP, Farzad R, Ha AD, Jivaji A, Karki S, Sheyn U, Stanton J, Minch B, Stephens D, Hancks DC, Rodrigues RAL, Abrahao JS, Vardi A, Aylward FO. Moniruzzaman M, et al. FEMS Microbiol Rev. 2023 Sep 5;47(5):fuad053. doi: 10.1093/femsre/fuad053. FEMS Microbiol Rev. 2023. PMID: 37740576 Free PMC article.
References
- Abergel, C., B. Coutard, D. Byrne, S. Chenivesse, J. B. Claude, C. Deregnaucourt, T. Fricaux, C. Gianesini-Boutreux, S. Jeudy, R. Lebrun, C. Maza, C. Notredame, O. Poirot, K. Suhre, M. Varagnol, and J.-M. Claverie. 2003. Structural genomics of highly conserved microbial genes of unknown function in search of new antibacterial targets. J. Struct. Funct. Genomics 4:141-157. - PubMed
- Adl, S. M., A. G. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, O. Moestrup, S. E. Mozley-Standridge, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. - PubMed
- Arnez, J. G., and D. Moras. 1997. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 22:211-216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials