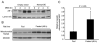Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival - PubMed (original) (raw)
. 2007 Sep 21;130(6):1095-107.
doi: 10.1016/j.cell.2007.07.035.
Tianle Yang, Joseph A Baur, Evelyn Perez, Takashi Matsui, Juan J Carmona, Dudley W Lamming, Nadja C Souza-Pinto, Vilhelm A Bohr, Anthony Rosenzweig, Rafael de Cabo, Anthony A Sauve, David A Sinclair
Affiliations
- PMID: 17889652
- PMCID: PMC3366687
- DOI: 10.1016/j.cell.2007.07.035
Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival
Hongying Yang et al. Cell. 2007.
Abstract
A major cause of cell death caused by genotoxic stress is thought to be due to the depletion of NAD(+) from the nucleus and the cytoplasm. Here we show that NAD(+) levels in mitochondria remain at physiological levels following genotoxic stress and can maintain cell viability even when nuclear and cytoplasmic pools of NAD(+) are depleted. Rodents fasted for 48 hr show increased levels of the NAD(+) biosynthetic enzyme Nampt and a concomitant increase in mitochondrial NAD(+). Increased Nampt provides protection against cell death and requires an intact mitochondrial NAD(+) salvage pathway as well as the mitochondrial NAD(+)-dependent deacetylases SIRT3 and SIRT4. We discuss the relevance of these findings to understanding how nutrition modulates physiology and to the evolution of apoptosis.
Figures
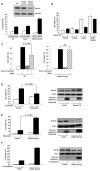
Figure 1. Nampt Is a Stress- and Nutrient-Responsive Gene that Protects Cells against the Genotoxic Agent MMS
(A and B) Nampt levels in human fibrosarcoma HT1080 cells in the presence or absence of 10% FBS (A) or of liver tissue extracts from fed or 2 day-fasted Sprague-Dawley rats (B). Actin and tubulin were used as loading controls. (C and D) Nampt protein (C) and mRNA levels of Nampt (D) in livers of fasted rats (n = 4; bars represent the mean of three experiments ± standard deviations [SD] using Student’s t test). (E) Western blot of Nampt in primary rat cardiomyocytes under hypoxia and/or serum starvation. (F and G) Survival of HT1080 cells stably expressing human (F) or transiently expressing mouse Nampt (G) following treatment with 1.2 mM methylmethanesulfonate (MMS). (H) Survival of human kidney HEK293 cells stably expressing human Nampt treated with MMS as in (F). Always, bars represent the mean of three experiments ± SD.
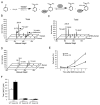
Figure 2. Nampt Protects against Apoptotic Cell Death Induced by Topoisomerase Inhibitors
(A) Sensitivity of HT1080 with siRNA-mediated knockdown of NAMPT after MMS exposure. (B) Stable overexpression of Nampt enhances survival of HEK293 cells following MMS treatment and the effect is blocked by the Nampt-inhibitor FK866. (C) Survival of WT or Nampt knockdown HT1080 cells after serum deprivation for 22 hr and then exposure to MMS for 17 hr. Serum deprivation upregulates Nampt and enhances survival of WT but not Nampt knockdown HT1080 cells. (D and E) Survival of HEK293 stably overexpressing Nampt (D) or HT1080 with siRNA knockdown of Nampt (E) following etoposide treatment. (F) Survival of HT1080 Nampt knockdown cells after camptothecin treatment. Apoptosis was assessed by western blot analysis of cleaved Caspase-3. Bars represent the mean of three experiments ± SD.
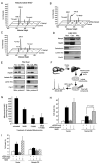
Figure 3. Nampt-Mediated Protection against Genotoxicity Requires SIRT3 and SIRT4
(A) Survival of HEK293 cells stably expressing Nampt following exposure to MMS in the presence or absence of the SIRT1-specific inhibitor EX-527. (B) siRNA knockdown of SIRT1 using a pool of four siRNA oligos, compared to nontargeting siRNA controls. Cells were cotransfected with FAM-tagged fluorescent oligos and percentage cell death was determined by FACS as a ratio of PI/FAM-positive cells versus total FAM-positive cells. (C) HEK293 cells were treated with 100 μM sirtinol, a pan sirtuin inhibitor. All experiments were carried out three times in triplicate. (D and E) SIRT3 or SIRT4 was knocked down in HEK293 cells stably overexpressing Nampt using pools of specific siRNA oligos, and cells were then treated with MMS and scored for survival. (F) Cells from (D) were probed by western blotting for cleaved caspase-3, an indicator of apoptosis. (G) Immunoprecipitation (IP) of AceCS2 from cell lysates of control and Nampt-overexpressing HEK293 cells transfected with control vector or AceCS2-HA for 48 hr. The levels of acetylated AceCS2 in IPs were analyzed by western blotting. Bars represent the mean of three experiments ± SD.
Figure 4. Nampt Regulates Total NAD+
(A) Synthesis of isotope-labeled 18O-NAD+, a reference compound used in NAD+ measurement. 18O-NAM was synthesized by hydrolyzing 3-cyanopyridine in 18O-H2O and was then used as a substrate in the enzymatic reaction catalyzed by CD38, a NAD+ glycohydrolase, to generate 18O-NAD+. (B and C) Total endogenous 16O-NAD+ and spiked-in NAD reference 18O-NAD+ were isolated by HPLC then subjected to MALDI-MS. The ion intensity of the reference peaks of 18O-NAD+ were normalized to 100 in all cases. The ratio of 16O-NAD+ peaks reflects the relative amount of NAD+ in the two samples. Experiments were performed at least three times. Total NAD+ spectra from HEK293 are shown for vector controls and cells stably overexpressing Nampt (B) as well as total NAD+ spectra from HT1080 vector controls and siRNA-Nampt stable cells (C). (D) Overexpression of Nampt cannot prevent total cellular NAD+ depletion by MMS as determined by MALDI-MS spectra of endogenous 16O-NAD and reference 18O-NAD after 2 hr MMS treatment of HEK293 WT and Nampt-overexpressing cells. (E) Time course of cell death induced by 1.2 mM MMS treatment. Percent cell death was determined by FACS analysis. (F) Total cellular NAD+ as measured by MALDI-MS during the time course in (E). Bars represent the mean of three experiments ± SD.
Figure 5. Mammalian Mitochondria Maintain Mitochondrial NAD+ Levels during Genotoxic Stress
(A and B) Nampt regulates mitochondrial NAD+ levels. Mitochondrial NAD+ was isolated and analyzed as described in Figures 4B and 4C. Spectra from HEK293 are shown for vector controls and cells stably overexpressing Nampt (A), as well as spectra from HT1080 vector controls and siRNA-Nampt stable cells (B). (C) Additional Nampt greatly attenuates mitochondrial NAD+ depletion by MMS treatment, as determined by MALDI-MS after 2 hr MMS treatment of HEK293 WT and Nampt-overexpressing cells. (D and E) Western blotting analysis of Nampt in highly purified cytosolic and mitochondrial fractions. Mitochondiral fractions were isolated from HEK293 cells or from rat livers using two different protocols, and their purity was assessed by probing for Hsp90, calreticulin, and/or lactate dehydrogenase (exclusively cytoplasmic proteins), and CoxIV or cytochrome C (mitochondrial matrix markers). The same blot was probed for lamin A/C to test for contamination of the mitochondrial fractions with nuclei. The experiment was performed three times on HEK293 cells and on liver tissue. The same pattern was observed each time and representative blots are shown. (F) Mitochondria from rat livers were prepared and exposed to methylmethane sulfonate (MMS), a genotoxic DNA alkylating agent, or the Nampt inhibitor FK866, or both. NAD+ levels in isolated mitochondria were determined using MALDI-MS, as above. (G) NAD+ levels in isolated mitochondria are reduced by exposure to MMS and FK866. Similar data were obtained using a different mitochondrial isolation protocol (see Figure S5). (H) Knocking down expression of Nmnat-3 reduces the ability of Nampt to provide resistance to MMS. (I) Knocking down expression of a putative human mitochondrial NAD+ transporter, hMFT, does not affect survival of Nampt-overexpressing cells treated with MMS. Bars represent the mean of three experiments ± SD.
Figure 6. Fasting Increases Hepatic Mitochondrial NAD+ and Nampt Levels
(A) Overexpression of Nampt in HEK293 cells inhibits the localization of AIF to the nucleus after MMS treatment for the times indicated, as assessed by western blotting. (B) Western blotting analysis of Nampt in mitochondria from rats fed AL or fasted for 48 hr. (C) Relative mitochondrial NAD+ levels in liver tissues from rats fed AL or fasted for 48 hr. Mitochondrial NAD+ levels were measured by MALDI-MS. Bars represent the mean of three experiments ± SD.
Similar articles
- Subcellular NAMPT-mediated NAD+ salvage pathways and their roles in bioenergetics and neuronal protection after ischemic injury.
Wang X, Zhang Z, Zhang N, Li H, Zhang L, Baines CP, Ding S. Wang X, et al. J Neurochem. 2019 Dec;151(6):732-748. doi: 10.1111/jnc.14878. Epub 2019 Oct 16. J Neurochem. 2019. PMID: 31553812 Free PMC article. - Mitochondrial sirtuins in the rat adrenal gland: location within the glands of males and females, hormonal and developmental regulation of gene expressions.
Celichowski P, Jopek K, Szyszka M, Tyczewska M, Malendowicz LK, Rucinski M. Celichowski P, et al. Folia Histochem Cytobiol. 2017;55(4):190-202. doi: 10.5603/FHC.a2017.0020. Epub 2017 Dec 20. Folia Histochem Cytobiol. 2017. PMID: 29261224 - Notoginseng Leaf Triterpenes Ameliorates OGD/R-Induced Neuronal Injury via SIRT1/2/3-Foxo3a-MnSOD/PGC-1_α_ Signaling Pathways Mediated by the NAMPT-NAD Pathway.
Xie W, Zhu T, Zhou P, Xu H, Meng X, Ding T, Nan F, Sun G, Sun X. Xie W, et al. Oxid Med Cell Longev. 2020 Oct 23;2020:7308386. doi: 10.1155/2020/7308386. eCollection 2020. Oxid Med Cell Longev. 2020. PMID: 33149812 Free PMC article. - Mitochondrial sirtuins.
Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Huang JY, et al. Biochim Biophys Acta. 2010 Aug;1804(8):1645-51. doi: 10.1016/j.bbapap.2009.12.021. Epub 2010 Jan 7. Biochim Biophys Acta. 2010. PMID: 20060508 Review. - Mitochondrial metabolism, sirtuins, and aging.
Sack MN, Finkel T. Sack MN, et al. Cold Spring Harb Perspect Biol. 2012 Dec 1;4(12):a013102. doi: 10.1101/cshperspect.a013102. Cold Spring Harb Perspect Biol. 2012. PMID: 23209156 Free PMC article. Review.
Cited by
- Intracellular nicotinamide adenine dinucleotide promotes TNF-induced necroptosis in a sirtuin-dependent manner.
Preyat N, Rossi M, Kers J, Chen L, Bertin J, Gough PJ, Le Moine A, Rongvaux A, Van Gool F, Leo O. Preyat N, et al. Cell Death Differ. 2016 Jan;23(1):29-40. doi: 10.1038/cdd.2015.60. Epub 2015 May 22. Cell Death Differ. 2016. PMID: 26001219 Free PMC article. - Tryptophan metabolism and disposition in cancer biology and immunotherapy.
Badawy AA. Badawy AA. Biosci Rep. 2022 Nov 30;42(11):BSR20221682. doi: 10.1042/BSR20221682. Biosci Rep. 2022. PMID: 36286592 Free PMC article. - Nicotinamide phosphoribosyl transferase (Nampt) is required for de novo lipogenesis in tumor cells.
Bowlby SC, Thomas MJ, D'Agostino RB Jr, Kridel SJ. Bowlby SC, et al. PLoS One. 2012;7(6):e40195. doi: 10.1371/journal.pone.0040195. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768255 Free PMC article. - NAMPT-Mediated NAD(+) Biosynthesis Is Essential for Vision In Mice.
Lin JB, Kubota S, Ban N, Yoshida M, Santeford A, Sene A, Nakamura R, Zapata N, Kubota M, Tsubota K, Yoshino J, Imai SI, Apte RS. Lin JB, et al. Cell Rep. 2016 Sep 27;17(1):69-85. doi: 10.1016/j.celrep.2016.08.073. Cell Rep. 2016. PMID: 27681422 Free PMC article. - Ceramide and mitochondria in ischemic brain injury.
Novgorodov SA, Gudz TI. Novgorodov SA, et al. Int J Biochem Mol Biol. 2011;2(4):347-61. Epub 2011 Nov 25. Int J Biochem Mol Biol. 2011. PMID: 22187669 Free PMC article.
References
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–18890. - PubMed
- Ando K, Higami Y, Tsuchiya T, Kanematsu T, Shimokawa I. Impact of aging and life-long calorie restriction on expression of apoptosis-related genes in male F344 rat liver. Microsc Res Tech. 2002;59:293–300. - PubMed
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AG027916-030003/AG/NIA NIH HHS/United States
- P01 AG027916-020003/AG/NIA NIH HHS/United States
- P01 AG027916-010003/AG/NIA NIH HHS/United States
- P01 AG027916-04S20003/AG/NIA NIH HHS/United States
- R01AG028730/AG/NIA NIH HHS/United States
- R01 AG019719-06A1/AG/NIA NIH HHS/United States
- R01 GM068072/GM/NIGMS NIH HHS/United States
- R01 AG028730/AG/NIA NIH HHS/United States
- R01 AG019719-07/AG/NIA NIH HHS/United States
- R01GM068072/GM/NIGMS NIH HHS/United States
- P01 AG027916-040003/AG/NIA NIH HHS/United States
- P01 AG027916/AG/NIA NIH HHS/United States
- P01 AG027916-04S10003/AG/NIA NIH HHS/United States
- R01 AG028730-02/AG/NIA NIH HHS/United States
- R01 DK073466/DK/NIDDK NIH HHS/United States
- R01 AG028730-01A1/AG/NIA NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 AG019719/AG/NIA NIH HHS/United States
- R01 DK 073466/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous