Family-based association tests for genomewide association scans - PubMed (original) (raw)
Family-based association tests for genomewide association scans
Wei-Min Chen et al. Am J Hum Genet. 2007 Nov.
Abstract
With millions of single-nucleotide polymorphisms (SNPs) identified and characterized, genomewide association studies have begun to identify susceptibility genes for complex traits and diseases. These studies involve the characterization and analysis of very-high-resolution SNP genotype data for hundreds or thousands of individuals. We describe a computationally efficient approach to testing association between SNPs and quantitative phenotypes, which can be applied to whole-genome association scans. In addition to observed genotypes, our approach allows estimation of missing genotypes, resulting in substantial increases in power when genotyping resources are limited. We estimate missing genotypes probabilistically using the Lander-Green or Elston-Stewart algorithms and combine high-resolution SNP genotypes for a subset of individuals in each pedigree with sparser marker data for the remaining individuals. We show that power is increased whenever phenotype information for ungenotyped individuals is included in analyses and that high-density genotyping of just three carefully selected individuals in a nuclear family can recover >90% of the information available if every individual were genotyped, for a fraction of the cost and experimental effort. To aid in study design, we evaluate the power of strategies that genotype different subsets of individuals in each pedigree and make recommendations about which individuals should be genotyped at a high density. To illustrate our method, we performed genomewide association analysis for 27 gene-expression phenotypes in 3-generation families (Centre d'Etude du Polymorphisme Humain pedigrees), in which genotypes for ~860,000 SNPs in 90 grandparents and parents are complemented by genotypes for ~6,700 SNPs in a total of 168 individuals. In addition to increasing the evidence of association at 15 previously identified cis-acting associated alleles, our genotype-inference algorithm allowed us to identify associated alleles at 4 cis-acting loci that were missed when analysis was restricted to individuals with the high-density SNP data. Our genotype-inference algorithm and the proposed association tests are implemented in software that is available for free.
Figures
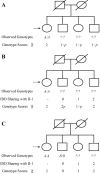
Figure 1.
Exemplar scoring of expected genotype scores. In each panel, the first sibling (individual II-1) is marked with an arrow. In panel A, only the first sibling is genotyped, and no flanking-marker data are available. In panel B, hypothetical flanking-marker data are available and can be used to characterize IBD sharing between the genotyped individual and her siblings. In panel C, two individuals are genotyped, providing further information.
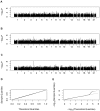
Figure 2.
Genome scan for CTBP1 expression levels. The gene maps to the beginning of chromosome 4. A, Genome scan using 60 unrelated individuals only. B, Genome scan using all 90 individuals genotyped by the HapMap Consortium. C, Genome scan that augmented the observed genotypes with expected genotype scores for other individuals, resulting in a total sample size of 156 individuals. All analysis were performed using the computationally efficient SCORE statistic. D, Q-Q plot. E, log Q–log Q plot. The plots show that the statistic is behaving adequately.
Similar articles
- In silico method for inferring genotypes in pedigrees.
Burdick JT, Chen WM, Abecasis GR, Cheung VG. Burdick JT, et al. Nat Genet. 2006 Sep;38(9):1002-4. doi: 10.1038/ng1863. Epub 2006 Aug 20. Nat Genet. 2006. PMID: 16921375 Free PMC article. - Characterisation of SNP haplotype structure in chemokine and chemokine receptor genes using CEPH pedigrees and statistical estimation.
Clark VJ, Dean M. Clark VJ, et al. Hum Genomics. 2004 Mar;1(3):195-207. doi: 10.1186/1479-7364-1-3-195. Hum Genomics. 2004. PMID: 15588479 Free PMC article. - Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis.
Aulchenko YS, de Koning DJ, Haley C. Aulchenko YS, et al. Genetics. 2007 Sep;177(1):577-85. doi: 10.1534/genetics.107.075614. Epub 2007 Jul 29. Genetics. 2007. PMID: 17660554 Free PMC article. - A 3.9-centimorgan-resolution human single-nucleotide polymorphism linkage map and screening set.
Matise TC, Sachidanandam R, Clark AG, Kruglyak L, Wijsman E, Kakol J, Buyske S, Chui B, Cohen P, de Toma C, Ehm M, Glanowski S, He C, Heil J, Markianos K, McMullen I, Pericak-Vance MA, Silbergleit A, Stein L, Wagner M, Wilson AF, Winick JD, Winn-Deen ES, Yamashiro CT, Cann HM, Lai E, Holden AL. Matise TC, et al. Am J Hum Genet. 2003 Aug;73(2):271-84. doi: 10.1086/377137. Epub 2003 Jul 3. Am J Hum Genet. 2003. PMID: 12844283 Free PMC article. - Recent developments in genomewide association scans: a workshop summary and review.
Thomas DC, Haile RW, Duggan D. Thomas DC, et al. Am J Hum Genet. 2005 Sep;77(3):337-45. doi: 10.1086/432962. Epub 2005 Aug 1. Am J Hum Genet. 2005. PMID: 16080110 Free PMC article. Review.
Cited by
- Unravelling the Genetic Architecture of Serum Biochemical Indicators in Sheep.
Kizilaslan M, Arzik Y, Behrem S, Yavuz E, White SN, Cinar MU. Kizilaslan M, et al. Genes (Basel). 2024 Jul 27;15(8):990. doi: 10.3390/genes15080990. Genes (Basel). 2024. PMID: 39202351 Free PMC article. - Genome-wide association analysis uncovers rice blast resistance alleles of Ptr and Pia.
Greenwood JR, Lacorte-Apostol V, Kroj T, Padilla J, Telebanco-Yanoria MJ, Glaus AN, Roulin A, Padilla A, Zhou B, Keller B, Krattinger SG. Greenwood JR, et al. Commun Biol. 2024 May 20;7(1):607. doi: 10.1038/s42003-024-06244-z. Commun Biol. 2024. PMID: 38769168 Free PMC article. - Genetic regions determine tolerance to nitrogen deficiency in European elite bread wheats grown under contrasting nitrogen stress scenarios.
Mini A, Touzy G, Beauchêne K, Cohan JP, Heumez E, Oury FX, Rincent R, Lafarge S, Le Gouis J; BreedWheat Consortium. Mini A, et al. Theor Appl Genet. 2023 Oct 10;136(11):218. doi: 10.1007/s00122-023-04468-x. Theor Appl Genet. 2023. PMID: 37815653 - Integrated epigenome, whole genome sequence and metabolome analyses identify novel multi-omics pathways in type 2 diabetes: a Middle Eastern study.
Yousri NA, Albagha OME, Hunt SC. Yousri NA, et al. BMC Med. 2023 Sep 8;21(1):347. doi: 10.1186/s12916-023-03027-x. BMC Med. 2023. PMID: 37679740 Free PMC article. - Symbiotic Variations among Wheat Genotypes and Detection of Quantitative Trait Loci for Molecular Interaction with Auxin-Producing Azospirillum PGPR.
Valente J, Gerin F, Mini A, Richard R, Le Gouis J, Prigent-Combaret C, Moënne-Loccoz Y. Valente J, et al. Microorganisms. 2023 Jun 19;11(6):1615. doi: 10.3390/microorganisms11061615. Microorganisms. 2023. PMID: 37375117 Free PMC article.
References
Web Resources
- Ghost, http://www.sph.umich.edu/csg/chen/ghost/ (for the Elston-Stewart–based implementation of our method)
- Merlin, http://www.sph.umich.edu/csg/abecasis/Merlin/ (for the Lander-Green–based implementation of our method)
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials