AAindex: amino acid index database, progress report 2008 - PubMed (original) (raw)
. 2008 Jan;36(Database issue):D202-5.
doi: 10.1093/nar/gkm998. Epub 2007 Nov 12.
Affiliations
- PMID: 17998252
- PMCID: PMC2238890
- DOI: 10.1093/nar/gkm998
AAindex: amino acid index database, progress report 2008
Shuichi Kawashima et al. Nucleic Acids Res. 2008 Jan.
Abstract
AAindex is a database of numerical indices representing various physicochemical and biochemical properties of amino acids and pairs of amino acids. We have added a collection of protein contact potentials to the AAindex as a new section. Accordingly AAindex consists of three sections now: AAindex1 for the amino acid index of 20 numerical values, AAindex2 for the amino acid substitution matrix and AAindex3 for the statistical protein contact potentials. All data are derived from published literature. The database can be accessed through the DBGET/LinkDB system at GenomeNet (http://www.genome.jp/dbget-bin/www\_bfind?aaindex) or downloaded by anonymous FTP (ftp://ftp.genome.jp/pub/db/community/aaindex/).
Figures
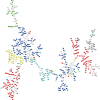
Figure 1.
The minimum spanning tree of the amino acid indices stored in the AAindex1 release 9.0. Each rectangle is an amino acid index. Colored nodes represent the indices classified by Tomii et al. (2) Red: alpha and turn propensities, Yellow: beta propensity, Green: composition, Blue: hydrophobicity, Cyan: physicochemical properties, Gray: other properties. White: the indices added to the AAindex after the release 3.0 by Tomii et al. (2).
Figure 2.
An example of database entry in the AAindex3. Each record of an entry is identified by the one-letter codes: H, accession number; D, definition of the entry; R, PMID identifier; A, author(s); T, title of the journal article; J, journal citation information; M, actual data in the specified order.
Similar articles
- AAindex: amino acid index database.
Kawashima S, Kanehisa M. Kawashima S, et al. Nucleic Acids Res. 2000 Jan 1;28(1):374. doi: 10.1093/nar/28.1.374. Nucleic Acids Res. 2000. PMID: 10592278 Free PMC article. - AAindex: Amino Acid Index Database.
Kawashima S, Ogata H, Kanehisa M. Kawashima S, et al. Nucleic Acids Res. 1999 Jan 1;27(1):368-9. doi: 10.1093/nar/27.1.368. Nucleic Acids Res. 1999. PMID: 9847231 Free PMC article. - LIGAND: database of chemical compounds and reactions in biological pathways.
Goto S, Okuno Y, Hattori M, Nishioka T, Kanehisa M. Goto S, et al. Nucleic Acids Res. 2002 Jan 1;30(1):402-4. doi: 10.1093/nar/30.1.402. Nucleic Acids Res. 2002. PMID: 11752349 Free PMC article. - DBGET/LinkDB: an integrated database retrieval system.
Fujibuchi W, Goto S, Migimatsu H, Uchiyama I, Ogiwara A, Akiyama Y, Kanehisa M. Fujibuchi W, et al. Pac Symp Biocomput. 1998:683-94. Pac Symp Biocomput. 1998. PMID: 9697222 - LIGAND: chemical database for enzyme reactions.
Goto S, Nishioka T, Kanehisa M. Goto S, et al. Bioinformatics. 1998;14(7):591-9. doi: 10.1093/bioinformatics/14.7.591. Bioinformatics. 1998. PMID: 9730924
Cited by
- FEGS: a novel feature extraction model for protein sequences and its applications.
Mu Z, Yu T, Liu X, Zheng H, Wei L, Liu J. Mu Z, et al. BMC Bioinformatics. 2021 Jun 3;22(1):297. doi: 10.1186/s12859-021-04223-3. BMC Bioinformatics. 2021. PMID: 34078264 Free PMC article. - PASE: a novel method for functional prediction of amino acid substitutions based on physicochemical properties.
Li X, Kierczak M, Shen X, Ahsan M, Carlborg O, Marklund S. Li X, et al. Front Genet. 2013 Mar 6;4:21. doi: 10.3389/fgene.2013.00021. eCollection 2013. Front Genet. 2013. PMID: 23508070 Free PMC article. - Prediction of anti-inflammatory peptides by a sequence-based stacking ensemble model named AIPStack.
Deng H, Lou C, Wu Z, Li W, Liu G, Tang Y. Deng H, et al. iScience. 2022 Aug 17;25(9):104967. doi: 10.1016/j.isci.2022.104967. eCollection 2022 Sep 16. iScience. 2022. PMID: 36093066 Free PMC article. - Sequence signatures of direct complementarity between mRNAs and cognate proteins on multiple levels.
Hlevnjak M, Polyansky AA, Zagrovic B. Hlevnjak M, et al. Nucleic Acids Res. 2012 Oct;40(18):8874-82. doi: 10.1093/nar/gks679. Epub 2012 Jul 25. Nucleic Acids Res. 2012. PMID: 22844092 Free PMC article. - On the Origin of Frameshift-Robustness of the Standard Genetic Code.
Xu H, Zhang J. Xu H, et al. Mol Biol Evol. 2021 Sep 27;38(10):4301-4309. doi: 10.1093/molbev/msab164. Mol Biol Evol. 2021. PMID: 34043802 Free PMC article.
References
- Nakai K, Kidera A, Kanehisa M. Cluster analysis of amino acid indices for prediction of protein structure and function. Protein Eng. 1988;2:93–100. - PubMed
- Tomii K, Kanehisa M. Analysis of amino acid indices and mutation matrices for sequence comparison and structure prediction of proteins. Protein Eng. 1996;9:27–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous