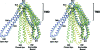Flexibility in the ABC transporter MsbA: Alternating access with a twist - PubMed (original) (raw)
Comparative Study
. 2007 Nov 27;104(48):19005-10.
doi: 10.1073/pnas.0709388104. Epub 2007 Nov 16.
Affiliations
- PMID: 18024585
- PMCID: PMC2141898
- DOI: 10.1073/pnas.0709388104
Comparative Study
Flexibility in the ABC transporter MsbA: Alternating access with a twist
Andrew Ward et al. Proc Natl Acad Sci U S A. 2007.
Abstract
ATP-binding cassette (ABC) transporters are integral membrane proteins that translocate a wide variety of substrates across cellular membranes and are conserved from bacteria to humans. Here we compare four x-ray structures of the bacterial ABC lipid flippase, MsbA, trapped in different conformations, two nucleotide-bound structures and two in the absence of nucleotide. Comparison of the nucleotide-free conformations of MsbA reveals a flexible hinge formed by extracellular loops 2 and 3. This hinge allows the nucleotide-binding domains to disassociate while the ATP-binding half sites remain facing each other. The binding of the nucleotide causes a packing rearrangement of the transmembrane helices and changes the accessibility of the transporter from cytoplasmic (inward) facing to extracellular (outward) facing. The inward and outward openings are mediated by two different sets of transmembrane helix interactions. Altogether, the conformational changes between these structures suggest that large ranges of motion may be required for substrate transport.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
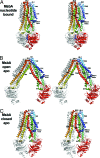
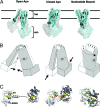
Fig. 1.
Stereoviews of three conformations of MsbA. (A) Nucleotide bound. (B) Open apo. (C) Closed apo. One monomer in each model is colored with a rainbow gradient (N terminus is blue, C terminus is red), and the other is white. TM helices (TM1–TM6), extracellular loops (EL1–EL3), and intracellular helices (IH1–IH2) are labeled accordingly. AMPPNP molecules are displayed as blue sticks in the nucleotide-bound structure. In all structures, TM4/TM5/IH2 (yellow and orange) associates with the opposite monomer in a conserved manner.
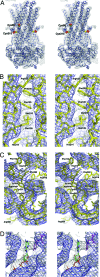
Fig. 2.
Electron-density maps for MsbA-AMPPNP. (A) Stereoview of a single MsbA-AMPPNP dimer (Cα model) contoured at 1σ (blue ribbon) with solvent-flattened and NCS-averaged density map (gray mesh) derived from MAD experimental phases. The heavy-atom mercury locations (red surface) and corresponding Cα positions of Cys-88 and -315 (yellow balls) are superimposed on the model. For clarity, the mercury positions have been labeled for only one monomer. The unaccounted density is from symmetry-related molecules. (B) Stereoview of side-chain electron density for MsbA-AMPPNP at 3.7-Å resolution derived from an averaging omit map phased from a model excluding nucleotide and truncated to polyalanine/glycine (white mesh, contour 1σ). The stacking of Phe-105 and -230 residues on adjacent TM helices and density for Arg-102, Met-109, Met-210, and His-214 are shown as yellow sticks. The initial phases for the averaging omit map were derived from a full model excluding nucleotide and truncated to polyalanine/glycine. The initial unaveraged map (blue mesh, contour 1σ) using polyalanine/glycine model phases is shown for reference. The recovered densities for the side chains are a consequence of the fourfold NCS averaging; these maps reveal the position of several bulky side chains and confirm the registration of amino acids in the model. (C) Stereoview of the same averaging omit map electron density (white mesh, contour 1σ) showing the positions of Phe-116, His-214, Phe-349, Tyr-351, Arg-360, Phe-392, Tyr-393, and Arg-416 in the NBD as yellow sticks. (D) Stereoview of averaging omit density (white mesh, contour 1σ) for AMPPNP (green, blue, red, orange sticks) and the MsbA-AMPPNP model (white sticks). Tyr-351 is shown as magenta sticks.
Fig. 3.
Electron-density maps for MsbA apo structures. (A) Stereoview of a single MsbA-open-apo dimer (Cα model) contoured at 1σ (blue ribbon) with solvent-flattened and NCS-averaged density map (gray mesh) derived from MAD/SIRAS combined experimental phases. (B) Stereoview of a single MsbA-closed-apo dimer (Cα model) contoured at 1σ (blue ribbon) with solvent-flattened and NCS-averaged density map (gray mesh) derived from MAD experimental phases. The heavy-atom Hg positions (red surface) and corresponding Cα positions (yellow balls) of Cys-88, -315, and -401 are superimposed onto the structure. For clarity, the mercury positions are labeled for only one monomer. The unaccounted density is from symmetry-related molecules.
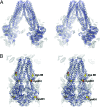
Fig. 4.
Comparison of the MsbA monomers. (A) Stereoview of the conformational changes within the MsbA monomer demonstrating the mobility of TM4/TM5/IH2. The NBDs have been removed for clarity. Monomers are aligned on TM helices 1, 2, 3, and 6. Two NCS-related monomers of MsbA-open-apo that differ in the relative positioning of TM4/TM5/IH2 are shown in blue and cyan (see also
SI Fig. 11
). The monomers from MsbA-closed-apo and MsbA-AMPPNP are shown in green and yellow, respectively. IH1 moves very little, whereas TM4/TM5/IH2 moves significantly.
Fig. 5.
Summary of conformational changes in MsbA. (A) Conformational changes within the MsbA dimer alter the accessibility to the internal chamber from inward to outward facing. For clarity, only TM helices (labeled 1–6) of one monomer (cyan) are shown inside a surface rendering of the dimer. The open and closed apo conformations form an inward-facing V between TM4/TM5 and TM3/TM6 (red asterisk). The nucleotide-bound conformation (MsbA-AMPPNP) forms an outward-facing V between TM3/TM6 and TM1/TM2, just above the elbow helix (black asterisk). Upon nucleotide binding, TM4/TM5/IH2 moves, causing TM3/TM6 to split away from TM1/TM2, which results in an outward-facing conformation. Both inward- and outward-facing conformations are mediated by intramolecular interactions within a single monomer, but by different sets of helices. (B) Simplified cartoon model illustrating the points above. The relative position of each TM helix is labeled with a number (one monomer in white and the other in gray). The arrows illustrate the motions required to go to the next state. (C) Top-down view of NBDs (one monomer shown in white and the other in gray). IH1 (green) and IH2 (yellow) from both monomers are shown. In the absence of nucleotide (apo), the NBDs are in similar orientations with the ATP-binding half-sites (LSGGQ and P-loop) facing each other; the P-loops (red) are roughly aligned (dashed lines) with one another across the dimer interface. Upon nucleotide binding (AMPPNP - magenta), the canonical ATP sandwich is formed, aligning the nucleotide between the LSGGQ and P-loop. IH1 tracks with the _cis_-monomer, whereas IH2 tracks with the _trans_-monomer. The motion of the NBDs from closed-apo- to nucleotide-bound transmits a structural change (described above) to the TMs via IH1 and IH2, resulting in an outward-facing conformation.
Similar articles
- Nucleotide dependent packing differences in helical crystals of the ABC transporter MsbA.
Ward A, Mulligan S, Carragher B, Chang G, Milligan RA. Ward A, et al. J Struct Biol. 2009 Mar;165(3):169-75. doi: 10.1016/j.jsb.2008.11.006. Epub 2008 Dec 10. J Struct Biol. 2009. PMID: 19114108 Free PMC article. - Structure of MsbA from Vibrio cholera: a multidrug resistance ABC transporter homolog in a closed conformation.
Chang G. Chang G. J Mol Biol. 2003 Jul 4;330(2):419-30. doi: 10.1016/s0022-2836(03)00587-4. J Mol Biol. 2003. PMID: 12823979 Retracted. - The conformational transition pathway of ATP binding cassette transporter MsbA revealed by atomistic simulations.
Weng JW, Fan KN, Wang WN. Weng JW, et al. J Biol Chem. 2010 Jan 29;285(5):3053-63. doi: 10.1074/jbc.M109.056432. Epub 2009 Dec 7. J Biol Chem. 2010. PMID: 19996093 Free PMC article. - Structure and mechanism of ABC transporter proteins.
Hollenstein K, Dawson RJ, Locher KP. Hollenstein K, et al. Curr Opin Struct Biol. 2007 Aug;17(4):412-8. doi: 10.1016/j.sbi.2007.07.003. Epub 2007 Aug 27. Curr Opin Struct Biol. 2007. PMID: 17723295 Review. - Investigating the dynamic nature of the ABC transporters: ABCB1 and MsbA as examples for the potential synergies of MD theory and EPR applications.
Stockner T, Mullen A, MacMillan F. Stockner T, et al. Biochem Soc Trans. 2015 Oct;43(5):1023-32. doi: 10.1042/BST20150138. Biochem Soc Trans. 2015. PMID: 26517918 Review.
Cited by
- Association/dissociation of the nucleotide-binding domains of the ATP-binding cassette protein MsbA measured during continuous hydrolysis.
Cooper RS, Altenberg GA. Cooper RS, et al. J Biol Chem. 2013 Jul 19;288(29):20785-20796. doi: 10.1074/jbc.M113.477976. Epub 2013 May 30. J Biol Chem. 2013. PMID: 23723071 Free PMC article. - In silico analysis of the binding of anthelmintics to Caenorhabditis elegansP-glycoprotein 1.
David MA, Orlowski S, Prichard RK, Hashem S, André F, Lespine A. David MA, et al. Int J Parasitol Drugs Drug Resist. 2016 Dec;6(3):299-313. doi: 10.1016/j.ijpddr.2016.09.001. Epub 2016 Sep 15. Int J Parasitol Drugs Drug Resist. 2016. PMID: 27746191 Free PMC article. - A Computational Approach towards the Understanding of Plasmodium falciparum Multidrug Resistance Protein 1.
Patel SK, George LB, Prasanth Kumar S, Highland HN, Jasrai YT, Pandya HA, Desai KR. Patel SK, et al. ISRN Bioinform. 2013 Aug 1;2013:437168. doi: 10.1155/2013/437168. eCollection 2013. ISRN Bioinform. 2013. PMID: 25937947 Free PMC article. - Nonequilibrium gating of CFTR on an equilibrium theme.
Jih KY, Hwang TC. Jih KY, et al. Physiology (Bethesda). 2012 Dec;27(6):351-61. doi: 10.1152/physiol.00026.2012. Physiology (Bethesda). 2012. PMID: 23223629 Free PMC article. Review. - Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans.
Jin MS, Oldham ML, Zhang Q, Chen J. Jin MS, et al. Nature. 2012 Oct 25;490(7421):566-9. doi: 10.1038/nature11448. Epub 2012 Sep 23. Nature. 2012. PMID: 23000902 Free PMC article.
References
- Higgins CF. Annu Rev Cell Biol. 1992;8:67–113. - PubMed
- Paulsen IT, Sliwinski MK, Saier MH. J Mol Biol. 1998;277:573–592. - PubMed
- Schneider E, Hunke S. FEMS Microbiol Rev. 1998;22:1–20. - PubMed
- Holland IB, Blight MA. J Mol Biol. 1999;293:381–399. - PubMed
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer J. Cell. 2000;101:789–800. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases