DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes - PubMed (original) (raw)
. 2008 Apr 1;22(7):908-17.
doi: 10.1101/gad.1640708.
Toshiaki Watanabe, Kengo Gotoh, Yasushi Totoki, Atsushi Toyoda, Masahito Ikawa, Noriko Asada, Kanako Kojima, Yuka Yamaguchi, Takashi W Ijiri, Kenichiro Hata, En Li, Yoichi Matsuda, Tohru Kimura, Masaru Okabe, Yoshiyuki Sakaki, Hiroyuki Sasaki, Toru Nakano
Affiliations
- PMID: 18381894
- PMCID: PMC2279202
- DOI: 10.1101/gad.1640708
DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes
Satomi Kuramochi-Miyagawa et al. Genes Dev. 2008.
Abstract
Silencing of transposable elements occurs during fetal gametogenesis in males via de novo DNA methylation of their regulatory regions. The loss of MILI (miwi-like) and MIWI2 (mouse piwi 2), two mouse homologs of Drosophila Piwi, activates retrotransposon gene expression by impairing DNA methylation in the regulatory regions of the retrotransposons. However, as it is unclear whether the defective DNA methylation in the mutants is due to the impairment of de novo DNA methylation, we analyze DNA methylation and Piwi-interacting small RNA (piRNA) expression in wild-type, MILI-null, and MIWI2-null male fetal germ cells. We reveal that defective DNA methylation of the regulatory regions of the Line-1 (long interspersed nuclear elements) and IAP (intracisternal A particle) retrotransposons in the MILI-null and MIWI2-null male germ cells takes place at the level of de novo methylation. Comprehensive analysis shows that the piRNAs of fetal germ cells are distinct from those previously identified in neonatal and adult germ cells. The expression of piRNAs is reduced under MILI- and MIWI2-null conditions in fetal germ cells, although the extent of the reduction differs significantly between the two mutants. Our data strongly suggest that MILI and MIWI2 play essential roles in establishing de novo DNA methylation of retrotransposons in fetal male germ cells.
Figures
Figure 1.
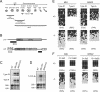
Expression of the IAP and Line-1 retrotransposons and methylation of their regulatory regions in neonatal prepachytene testes. (A) Scheme for the development of mouse male germ cells. (Gonia) Spermatogonia; (Lep) leptotene; (Zy) zygotene; (Pachy) pachytene; (RSp) round spermatid; (Esp) elongated spermatid. (B) Schematic diagram of the Line-1 and IAP genes. The locations of the probes used for Northern blotting and bisulfite sequencing are indicated by filled and open boxes, respectively. The sequences of the 5′-noncoding regions of the Line-1 genes are different for type Gf and type A. The probe for the 3′-noncoding region of IAP recognizes the full-length and all deletion derivatives of IAP. (C,D) Northern blotting showing transcription of the Line-1 (C) and IAP (D) retrotransposon genes in testes from 2-wk-old MILI- and MIWI2-deficient mice. The 5′-noncoding regions of type Gf and type A Line-1 and the 3′-noncoding region of IAP were used as probes. (E,F) Bisulfite sequencing of Line-1 (E) and IAP (F) in MILI- and MIWI2-deficient germ cells. Spermatogonial cells from 6- to 12-d-old mice were purified from Oct4-EGFP transgenic mouse (Yoshimizu et al. 1999) testes. (E) The 5′-noncoding regions of type Gf and type A Line-1 (nucleotides 874–1156; GenBank accession no. D84391) and nucleotides 1251–1542 of M13002 were analyzed. (F) Two LTR regions from the 5.4-kb IΔ1-type IAP in chromosomes 3qD and 16qB2 were arbitrarily chosen for analysis. Filled and open circles represent methylated and unmethylated CpGs, respectively. The percentage of methylated CpGs is shown in parentheses.
Figure 2.
Methylation of the IAP and Line-1 regulatory regions in fetal testes. (A) Methylation-sensitive Southern blot analysis of the Line-1 promoter region. Whole-testis DNA was extracted from 2-d-old heterozygous (+) and homozygous (−) mice, followed by digestion with KpnI and the methylation-sensitive restriction enzyme HpaII. The probe for the type Gf Line-1 5′-noncoding region is the same as that in Figure 1C. (B) Bisulfite analysis of the Line-1 regulatory region in day 0–1 germ cells from MILI- and MIWI2-deficient and control testes. The germ cells were collected as described in Materials and Methods. (C,D) Bisulfite sequencing of the Line-1 (C) and IAP (D) regulatory regions in germ cells from MILI-deficient and control testes between E13.5 and E18.5. The germ cells were collected as described in Materials and Methods.
Figure 3.
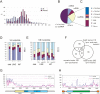
piRNAs in fetal premeiotic germ cells. (A) In total, 127,997 small RNAs were sequenced from E12.5–E19.5 fetal germ cells. The size distributions (in nucleotides) of the total small RNAs and rasiRNAs are shown by blue and purple bars, respectively. (B,C) Genomic annotation of the small RNAs (B) and the ratio of piRNA sequences of fetal premeiotic germ cells (C). Detailed results are listed in Supplemental Table S2. (D,E) Comparison of the first (D) and tenth (E) nucleotides of the total, sense (+), and antisense (−) piRNAs. Nucleotide biases were calculated for the Gf type Line-1 and IAP piRNAs analyzed in Supplemental Table S2. (E) The piRNA classes that contain or lack a 5′-end U are shown separately. (F) Venn diagram of the piRNAs in adult pre- and post-meiotic (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2006), neonatal prepachytene (Aravin et al. 2007), and fetal (this study) testes. (G,H) Distribution of piRNAs corresponding to type Gf Line-1 (G) and IAPIΔ1 (H).
Figure 4.
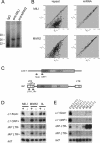
piRNA expression in MILI- and MIWI2-deficient fetal testes. (A) MILI- and MIWI2-bound piRNAs. The immunoprecipitated RNAs from E16.5 testicular lysates were 32P-end-labeled and separated in a 15% denaturing urea–polyacrylamide gel. (B) Microarray analysis of the repeat-associated piRNAs and miRNAs. A microarray that contained 672 types of repeat-associated piRNAs and 150 types of miRNAs, both of which are expressed in fetal testes, was produced. RNA samples isolated from E16.5 control, MILI-deficient, and MIWI2-deficient mice were used. The expression of each small RNA was examined three times, and the mean values are plotted to compare the control and MILI- or MIWI2-deficient samples. Diagonal lines indicate a 1.5-fold difference in expression. (C–E) Expression of four arbitrarily selected piRNAs. piRNAs corresponding to the type A Line-1 promoter sense strand (5UA+), Line-1 ORF sense strand (ORF+), IAP LTR sense strand (LTR+), and IAP LTR antisense strand (LTR−) were arbitrarily selected. (C) The site and orientation of each piRNA is indicated by red arrows. (D) Northern blot analysis of the piRNAs in E16.5 control, MILI-deficient, MIWI2-deficient, and Dnmt3L-deficient testis. (E) Time-course analysis of the expression of the four piRNAs in control and MILI-null testes.
Comment in
- Small RNA guides for de novo DNA methylation in mammalian germ cells.
Aravin AA, Bourc'his D. Aravin AA, et al. Genes Dev. 2008 Apr 15;22(8):970-5. doi: 10.1101/gad.1669408. Genes Dev. 2008. PMID: 18413711 Free PMC article.
Similar articles
- MIWI2 as an Effector of DNA Methylation and Gene Silencing in Embryonic Male Germ Cells.
Kojima-Kita K, Kuramochi-Miyagawa S, Nagamori I, Ogonuki N, Ogura A, Hasuwa H, Akazawa T, Inoue N, Nakano T. Kojima-Kita K, et al. Cell Rep. 2016 Sep 13;16(11):2819-2828. doi: 10.1016/j.celrep.2016.08.027. Cell Rep. 2016. PMID: 27626653 - MVH in piRNA processing and gene silencing of retrotransposons.
Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, Totoki Y, Shibata T, Kimura T, Nakatsuji N, Noce T, Sasaki H, Nakano T. Kuramochi-Miyagawa S, et al. Genes Dev. 2010 May;24(9):887-92. doi: 10.1101/gad.1902110. Genes Dev. 2010. PMID: 20439430 Free PMC article. - The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements.
De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. De Fazio S, et al. Nature. 2011 Oct 23;480(7376):259-63. doi: 10.1038/nature10547. Nature. 2011. PMID: 22020280 - Small RNA silencing pathways in germ and stem cells.
Aravin AA, Hannon GJ. Aravin AA, et al. Cold Spring Harb Symp Quant Biol. 2008;73:283-90. doi: 10.1101/sqb.2008.73.058. Epub 2009 Mar 6. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19270082 Review. - Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline.
Crichton JH, Dunican DS, Maclennan M, Meehan RR, Adams IR. Crichton JH, et al. Cell Mol Life Sci. 2014 May;71(9):1581-605. doi: 10.1007/s00018-013-1468-0. Epub 2013 Sep 18. Cell Mol Life Sci. 2014. PMID: 24045705 Free PMC article. Review.
Cited by
- Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology.
Martinez VD, Vucic EA, Thu KL, Hubaux R, Enfield KS, Pikor LA, Becker-Santos DD, Brown CJ, Lam S, Lam WL. Martinez VD, et al. Sci Rep. 2015 May 27;5:10423. doi: 10.1038/srep10423. Sci Rep. 2015. PMID: 26013764 Free PMC article. - The copy number of rice CACTA DNA transposons carrying MIR820 does not correlate with MIR820 expression.
Nosaka M, Ishiwata A, Shimizu-Sato S, Ono A, Ishimoto K, Noda Y, Sato Y. Nosaka M, et al. Plant Signal Behav. 2013 Aug;8(8):e25169. doi: 10.4161/psb.25169. Epub 2013 May 31. Plant Signal Behav. 2013. PMID: 23733074 Free PMC article. - Mutations in the MOV10L1 ATP Hydrolysis Motif Cause piRNA Biogenesis Failure and Male Sterility in Mice.
Fu Q, Pandey RR, Leu NA, Pillai RS, Wang PJ. Fu Q, et al. Biol Reprod. 2016 Nov 1;95(5):103. doi: 10.1095/biolreprod.116.142430. Epub 2016 Sep 21. Biol Reprod. 2016. PMID: 27655786 Free PMC article. - NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing.
Shirane K, Miura F, Ito T, Lorincz MC. Shirane K, et al. Nat Genet. 2020 Oct;52(10):1088-1098. doi: 10.1038/s41588-020-0689-z. Epub 2020 Sep 14. Nat Genet. 2020. PMID: 32929285 - Endogenous Retroviruses Walk a Fine Line between Priming and Silencing.
Cullen H, Schorn AJ. Cullen H, et al. Viruses. 2020 Jul 23;12(8):792. doi: 10.3390/v12080792. Viruses. 2020. PMID: 32718022 Free PMC article. Review.
References
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. - PubMed
- Bourc’his D., Bestor T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. - PubMed
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials