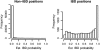Estimation of pairwise identity by descent from dense genetic marker data in a population sample of haplotypes - PubMed (original) (raw)
Estimation of pairwise identity by descent from dense genetic marker data in a population sample of haplotypes
Sharon R Browning. Genetics. 2008 Apr.
Abstract
I present a new approach for calculating probabilities of identity by descent for pairs of haplotypes. The approach is based on a joint hidden Markov model for haplotype frequencies and identity by descent (IBD). This model allows for linkage disequilibrium, and the method can be applied to very dense marker data. The method has high power for detecting IBD tracts of genetic length of 1 cM, with the use of sufficiently dense markers. This enables detection of pairwise IBD between haplotypes from individuals whose most recent common ancestor lived up to 50 generations ago.
Figures
Figure 1.—
Example of a localized haplotype cluster model. Nodes of the graph represent haplotype clusters (labeled A–L). Solid boundaries around nodes represent allele 1 while dashed boundaries represent allele 2. The initial state is shaded gray. Transition probabilities are given on the edges.
Figure 2.—
An alternative representation of the localized haplotype cluster model in Figure 1. This representation is given solely for comparison with previous work (for example, B
rowning
2006). Edges of the graph represent the haplotype clusters. Solid edges represent allele 1 while dashed edges represent allele 2.
Figure 3.—
Estimated IBD probabilities. Histograms of maximum estimated IBD probabilities from each region. (Top) Results from pairs of haplotypes with no IBD. (Middle) Results from pairs of haplotypes with short tracts of IBD. (Bottom) Results from pairs of haplotypes with long tracts of IBD. The genetic lengths given are approximate.
Figure 4.—
Estimated IBD probabilities for random IBD data. IBD tracts were inserted in pairs of simulated haplotypes at random according to the prior distribution. IBD probabilities were estimated at both IBD and non-IBD positions, and these probabilities are shown in the two histograms.
Figure 5.—
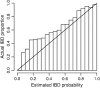
Accuracy of estimated IBD probabilities. IBD tracts were inserted in pairs of simulated haplotypes at random according to the prior distribution. IBD probabilities were estimated at both IBD and non-IBD positions. Within each bin shown on the _x_-axis, the locations having an estimated IBD probability falling within the bin are considered. Of those locations falling into the bin, the proportion of locations that are actually part of an IBD tract is shown by the height of the bar for that bin. As the distribution of IBD follows the same distribution as the prior used in the IBD estimation, actual proportions should ideally match the estimated probabilities and thus fall close to the diagonal line.
Similar articles
- Optimal haplotype structure for linkage disequilibrium-based fine mapping of quantitative trait loci using identity by descent.
Grapes L, Firat MZ, Dekkers JC, Rothschild MF, Fernando RL. Grapes L, et al. Genetics. 2006 Mar;172(3):1955-65. doi: 10.1534/genetics.105.048686. Epub 2005 Dec 1. Genetics. 2006. PMID: 16322505 Free PMC article. - Multipoint identity-by-descent prediction using dense markers to map quantitative trait loci and estimate effective population size.
Meuwissen TH, Goddard ME. Meuwissen TH, et al. Genetics. 2007 Aug;176(4):2551-60. doi: 10.1534/genetics.107.070953. Epub 2007 Jun 11. Genetics. 2007. PMID: 17565953 Free PMC article. - The probability that similar haplotypes are identical by descent.
Nolte IM, Te Meerman GJ. Nolte IM, et al. Ann Hum Genet. 2002 May;66(Pt 3):195-209. doi: 10.1017/S0003480002001100. Ann Hum Genet. 2002. PMID: 12174211 - Estimating genealogies from unlinked marker data: a Bayesian approach.
Gasbarra D, Pirinen M, Sillanpää MJ, Salmela E, Arjas E. Gasbarra D, et al. Theor Popul Biol. 2007 Nov;72(3):305-22. doi: 10.1016/j.tpb.2007.06.004. Epub 2007 Jun 22. Theor Popul Biol. 2007. PMID: 17681576 Review. - [Genetic aspects of genealogy].
Tetushkin EIu. Tetushkin EIu. Genetika. 2011 Nov;47(11):1451-72. Genetika. 2011. PMID: 22332404 Review. Russian.
Cited by
- The influence of gene flow on population viability in an isolated urban caracal population.
Kyriazis CC, Serieys LEK, Bishop JM, Drouilly M, Viljoen S, Wayne RK, Lohmueller KE. Kyriazis CC, et al. Mol Ecol. 2024 May;33(9):e17346. doi: 10.1111/mec.17346. Epub 2024 Apr 5. Mol Ecol. 2024. PMID: 38581173 - Biobank-scale inference of multi-individual identity by descent and gene conversion.
Browning SR, Browning BL. Browning SR, et al. Am J Hum Genet. 2024 Apr 4;111(4):691-700. doi: 10.1016/j.ajhg.2024.02.015. Epub 2024 Mar 20. Am J Hum Genet. 2024. PMID: 38513668 Free PMC article. - Biobank-scale inference of multi-individual identity by descent and gene conversion.
Browning SR, Browning BL. Browning SR, et al. bioRxiv [Preprint]. 2023 Nov 5:2023.11.03.565574. doi: 10.1101/2023.11.03.565574. bioRxiv. 2023. PMID: 37961601 Free PMC article. Updated. Preprint. - IBD sharing patterns as intra-breed admixture indicators in small ruminants.
Blondeau Da Silva S, Mwacharo JM, Li M, Ahbara A, Muchadeyi FC, Dzomba EF, Lenstra JA, Da Silva A. Blondeau Da Silva S, et al. Heredity (Edinb). 2024 Jan;132(1):30-42. doi: 10.1038/s41437-023-00658-x. Epub 2023 Nov 3. Heredity (Edinb). 2024. PMID: 37919398 Free PMC article. - The genomic footprint of whaling and isolation in fin whale populations.
Nigenda-Morales SF, Lin M, Nuñez-Valencia PG, Kyriazis CC, Beichman AC, Robinson JA, Ragsdale AP, Urbán R J, Archer FI, Viloria-Gómora L, Pérez-Álvarez MJ, Poulin E, Lohmueller KE, Moreno-Estrada A, Wayne RK. Nigenda-Morales SF, et al. Nat Commun. 2023 Sep 12;14(1):5465. doi: 10.1038/s41467-023-40052-z. Nat Commun. 2023. PMID: 37699896 Free PMC article.
References
- Baum, L. E., 1972. An inequality and associated maximization technique in statistical estimation for probabilistic functions on Markov processes, pp. 1–8 in Inequalities. III. Proceedings of the Third Symposium on Inequalities. Academic Press, New York.
- Beckmann, L., D. C. Thomas, C. Fischer and J. Chang-Claude, 2005. Haplotype sharing analysis using Mantel statistics. Hum. Hered. 59 67–78. - PubMed
- Browning, B. L., and S. R. Browning, 2007. Efficient multilocus association mapping for whole genome association studies using localized haplotype clustering. Genet. Epidemiol. 31 365–375. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources