Glucose-mediated transactivation of carbohydrate response element-binding protein requires cooperative actions from Mondo conserved regions and essential trans-acting factor 14-3-3 - PubMed (original) (raw)
Glucose-mediated transactivation of carbohydrate response element-binding protein requires cooperative actions from Mondo conserved regions and essential trans-acting factor 14-3-3
Ming V Li et al. Mol Endocrinol. 2008 Jul.
Abstract
Carbohydrate response element-binding protein (ChREBP) is a basic helix-loop-helix/leucine zipper transcription factor that binds to the carbohydrate response element in the promoter of certain lipogenic and glycolytic genes. High glucose can activate ChREBP by releasing an intramolecular inhibition within the glucose-sensing module (GSM) that occurs in low glucose. We report here that the glucose response of GSM is mediated by cooperation between five conserved submodules known as Mondo conserved regions (MCRs) I through V within GSM. Deletion of individual MCRs leads to complete (for MCR II, III, and IV) or partial (MCR I) loss of glucose response of ChREBP. MCR IV is necessary and sufficient for inhibiting the transcriptional activity of ChREBP under low glucose. The roles of MCR II and III in glucose response of ChREBP are independent of and distinct from their function in controlling subcellular localization. We further demonstrate that, instead of inhibiting ChREBP activity as would be predicted from its cytoplasmic retentive function, 14-3-3 binding with MCR III is essential for the glucose responsiveness of ChREBP. The interaction between 14-3-3 and ChREBP is constitutive, indicating a permissive role of 14-3-3 in the glucose response of ChREBP. We further uncovered an unconventional 14-3-3 binding motif (residues 116-135) lacking phosphor-serine/threonine within MCR III, a predicted alpha-helix highly conserved in all Mondo proteins. We conclude that individual subdomains in the GSM (MCR I through V) play diverse but crucial roles in cooperation with essential trans-acting cofactors such as 14-3-3 proteins to mediate the glucose response of ChREBP.
Figures
Figure 1
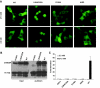
Structure-Function Analysis of the GSM A, Schematic illustration of the deletion constructs (not to scale). Each MCR is labeled as I through V, and regions corresponding to LID and GRACE are bracketed. B, Expression of above constructs after being transfected into 832/13 cells for 24 h as examined by Western blot using anti-c-myc antibody. C, Luciferase assay. pG5-luc and pRL-TK were cotransfected with indicated plasmids into 832/13 cells. Transfected cells were treated with 2.5 m
m
(white bar) or 27.5 m
m
(black bar) glucose for 24 h.
Figure 2
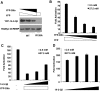
Subcellular Localization and Glucose Response of ChREBP and Its Mutants A, Immunofluorescent staining. The 832/13 cells stably expressing c-_myc_-tagged wild-type or indicated mutant ChREBP were treated with low (2.5 m
m
) or high (27.5 m
m
) glucose for 12 h before staining with anti-c-myc antibody. B, Poly-His pull-down assay. pCHM-14-3-3β was cotransfected with plasmids expressing indicated wild-type or mutant ChREBP into 832/13 cells. Twenty-four hours after transfection, we used Western blot with anti-c-myc antibody to examine the levels of expressed proteins in the cell lysate (input) or in the precipitate after pull-down assay. C, Luciferase assay. pG5-luc and pRL-TK were cotransfected with indicated plasmids into 832/13 cells that were treated with 2.5 or 27.5 m
m
glucose for 24 h. WT, Wild type.
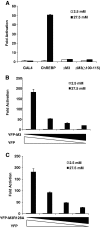
Figure 3
14-3-3 Proteins Are Essential for Glucose Response of ChREBP A, Poly-His pull-down assay. pCHM-ChREBP or its R128A mutant, pYFP-14-3-3β and different doses of pYFP and pYFP-difo as indicated, were transfected into 832/13 cells. We collected the cell lysate after 24 h and performed poly-His pull-down assay. The amount of YFP-14-3-3β and ChREBP pulled down were measured with Western blot using anti-GFP and anti-c-myc antibodies, respectively. B–D, Luciferase assays. B, pG5-luc, pRL-TK, and pGAMPAC-ChREBP were cotransfected with indicated amounts of pYFP-Difo or pYFP into 832/13 cells that were treated with 2.5 or 27.5 m
m
glucose for 24 h. C, pG5-luc and pRL-TK were cotransfected with plasmids expressing indicated proteins into 832/13 cells that were treated with 2.5 or 27.5 m
m
glucose for 24 h. The total amounts of transfected DNA for each group were held the same with stuffer plasmid pcDNA3.1. D, pG5-luc and pRL-TK were cotransfected with different amounts of pCHM-14-3-3β into 832/13 cells that were treated with 2.5 or 27.5 m
m
glucose for 24 h. The total amounts of transfected DNA for each group were held the same with stuffer plasmid pcDNA3.1. WT, Wild type.
Figure 4
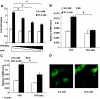
The 14-3-3 Proteins Are Required for Glucose-Stimulated Expression of ChREBP Target Genes A, Luciferase assay. pLPK-luc and pRL-TK were cotransfected with the indicated amounts of pYFP-Difo or pYFP into 832/13 cells that were treated with 2.5 or 27.5 m
m
glucose for 24 h. B and C, Quantitative RT-PCR. The 832/13 cells overexpressing YFP or YFP-Difo were treated with 2.5 or 27.5 m
m
glucose for 48 h. The levels of FAS (B) and ACC (C) were measured with real-time RT-PCR. *, P < 0.01. D, Subcellular localization of YFP-14-3-3β fusion protein. The 832/13 cells were infected with retrovirus expressing YFP-14-3-3β fusion protein and treated with 2.5 or 27.5 m
m
glucose for 12 h.
Figure 5
Characterization of Interaction between ChREBP and 14-3-3 A and B, Poly-His pull-down assay. The 832/13 cells stably expressing 6×His/c-_myc_-tagged 14-3-3β and c-_myc_-tagged ChREBP or its R128 mutant were treated with low (L, 2.5 m
m
) or high (H, 27.5 m
m
) glucose for 4 h and then either directly lysed for the pull-down assay (A) or treated with DSP before lysis for pull-down assay (B). C, Poly-His pull-down assay. pCHM-14-3-3β was cotransfected with plasmids expressing indicated wild-type or mutant ChREBP into 832/13 cells. Cells were lysed after 24 h for poly-His pull-down assay. D, GST pull-down assay. Glutathione-Sepharose beads coated with the indicated recombinant protein were incubated with recombinant c-_myc_-tagged 14-3-3β. The amount of 14-3-3β retained on these beads was examined by Western blot with anti-c-myc antibody. WT, Wild type.
Figure 6
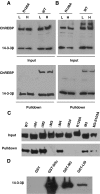
Identification of the 14-3-3-Interacting Motif in ChREBP A, Secondary structure prediction with SSpro8 program. Key residues were marked by asterisks and their number in the ChREBP amino acid sequence. The conserved α-helix (116–135) is boxed. Codes for secondary structure: C, coil; H, α-helix; T, turn; E, extended strand. B, Schematic illustration of the ChREBP deletion constructs (not to scale). C, Poly-His pull-down assay. pCHM-14-3-3β was cotransfected with indicated plasmids shown in B into 832/13 cells. Cells were lysed 24 h after transfection for poly-His pull-down assay.
Figure 7
The 14-3-3-Independent Function of MCR III A–C, Luciferase assay: A, pG5-luc and pRL-TK were cotransfected with plasmids expressing indicated proteins into 832/13 cells that were treated with 2.5 or 27.5 m
m
glucose for 24 h; B, pG5-luc, pRL-TK and pGAMPAC-ChREBP were cotransfected with different amounts of pYFP-M3 or pYFP into 832/13 cells that were treated with 2.5 or 27.5 m
m
glucose for 24 h. C, pG5-luc, pRL-TK, and pGAMPAC-ChREBP were cotransfected with different amounts of pYFP-M3R128A or pYFP into 832/13 cells that were treated with 2.5 or 27.5 m
m
glucose for 24 h.
Similar articles
- Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module.
Li MV, Chang B, Imamura M, Poungvarin N, Chan L. Li MV, et al. Diabetes. 2006 May;55(5):1179-89. doi: 10.2337/db05-0822. Diabetes. 2006. PMID: 16644671 - Activation and repression of glucose-stimulated ChREBP requires the concerted action of multiple domains within the MondoA conserved region.
Davies MN, O'Callaghan BL, Towle HC. Davies MN, et al. Am J Physiol Endocrinol Metab. 2010 Oct;299(4):E665-74. doi: 10.1152/ajpendo.00349.2010. Epub 2010 Aug 3. Am J Physiol Endocrinol Metab. 2010. PMID: 20682844 Free PMC article. - A novel N-terminal domain may dictate the glucose response of Mondo proteins.
McFerrin LG, Atchley WR. McFerrin LG, et al. PLoS One. 2012;7(4):e34803. doi: 10.1371/journal.pone.0034803. Epub 2012 Apr 10. PLoS One. 2012. PMID: 22506051 Free PMC article. - The physiological and pathophysiological roles of carbohydrate response element binding protein in the kidney.
Yokoyama A, Suzuki S, Okamoto K, Sugawara A. Yokoyama A, et al. Endocr J. 2022 Jun 28;69(6):605-612. doi: 10.1507/endocrj.EJ22-0083. Epub 2022 Apr 26. Endocr J. 2022. PMID: 35474028 Review. - The Roles of Carbohydrate Response Element Binding Protein in the Relationship between Carbohydrate Intake and Diseases.
Iizuka K. Iizuka K. Int J Mol Sci. 2021 Nov 8;22(21):12058. doi: 10.3390/ijms222112058. Int J Mol Sci. 2021. PMID: 34769488 Free PMC article. Review.
Cited by
- Glucose induces protein targeting to glycogen in hepatocytes by fructose 2,6-bisphosphate-mediated recruitment of MondoA to the promoter.
Petrie JL, Al-Oanzi ZH, Arden C, Tudhope SJ, Mann J, Kieswich J, Yaqoob MM, Towle HC, Agius L. Petrie JL, et al. Mol Cell Biol. 2013 Feb;33(4):725-38. doi: 10.1128/MCB.01576-12. Epub 2012 Dec 3. Mol Cell Biol. 2013. PMID: 23207906 Free PMC article. - O-GlcNAcylation Links ChREBP and FXR to Glucose-Sensing.
Benhamed F, Filhoulaud G, Caron S, Lefebvre P, Staels B, Postic C. Benhamed F, et al. Front Endocrinol (Lausanne). 2015 Jan 13;5:230. doi: 10.3389/fendo.2014.00230. eCollection 2014. Front Endocrinol (Lausanne). 2015. PMID: 25628602 Free PMC article. Review. - Genome-Wide Analysis of ChREBP Binding Sites on Male Mouse Liver and White Adipose Chromatin.
Poungvarin N, Chang B, Imamura M, Chen J, Moolsuwan K, Sae-Lee C, Li W, Chan L. Poungvarin N, et al. Endocrinology. 2015 Jun;156(6):1982-94. doi: 10.1210/en.2014-1666. Epub 2015 Mar 9. Endocrinology. 2015. PMID: 25751637 Free PMC article. - The glucose-sensing transcription factor ChREBP is targeted by proline hydroxylation.
Heidenreich S, Weber P, Stephanowitz H, Petricek KM, Schütte T, Oster M, Salo AM, Knauer M, Goehring I, Yang N, Witte N, Schumann A, Sommerfeld M, Muenzner M, Myllyharju J, Krause E, Schupp M. Heidenreich S, et al. J Biol Chem. 2020 Dec 11;295(50):17158-17168. doi: 10.1074/jbc.RA120.014402. Epub 2020 Oct 6. J Biol Chem. 2020. PMID: 33023907 Free PMC article. - The Krüppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription.
Yang Y, Chang BH, Samson SL, Li MV, Chan L. Yang Y, et al. Nucleic Acids Res. 2009 May;37(8):2529-38. doi: 10.1093/nar/gkp122. Epub 2009 Mar 5. Nucleic Acids Res. 2009. PMID: 19264802 Free PMC article.
References
- Thompson KS, Towle HC 1991 Localization of the carbohydrate response element of the rat L-type pyruvate kinase gene. J Biol Chem 266:8679–8682 - PubMed