A generic approach for the purification of signaling complexes that specifically interact with the carboxyl-terminal domain of G protein-coupled receptors - PubMed (original) (raw)
doi: 10.1074/mcp.M700435-MCP200. Epub 2008 Apr 29.
Avais M Daulat, Cédric Broussard, Julien Mozo, Guilhem Clary, Françoise Hotellier, Philippe Chafey, Jean-Luc Guillaume, Gilles Ferry, Jean A Boutin, Philippe Delagrange, Luc Camoin, Ralf Jockers
Affiliations
- PMID: 18448421
- PMCID: PMC2500234
- DOI: 10.1074/mcp.M700435-MCP200
A generic approach for the purification of signaling complexes that specifically interact with the carboxyl-terminal domain of G protein-coupled receptors
Pascal Maurice et al. Mol Cell Proteomics. 2008 Aug.
Abstract
G protein-coupled receptors (GPCRs) constitute the largest family of membrane receptors and are major drug targets. Recent progress has shown that GPCRs are part of large protein complexes that regulate their activity. We present here a generic approach for identification of these complexes that is based on the use of receptor subdomains and that overcomes the limitations of currently used genetics and proteomics approaches. Our approach consists of a carefully balanced combination of chemically synthesized His6-tagged baits, immobilized metal affinity chromatography, one- and two-dimensional gel electrophoresis separation and mass spectrometric identification. The carboxyl-terminal tails (C-tails) of the human MT1 and MT2 melatonin receptors, two class A GPCRs, were used as models to purify protein complexes from mouse brain lysates. We identified 32 proteins that interacted with the C-tail of MT1, 14 proteins that interacted with the C-tail of MT2, and eight proteins that interacted with both C-tails. Several randomly selected proteins were validated by Western blotting, and the functional relevance of our data was further confirmed by showing the interaction between the full-length MT1 and the regulator of G protein signaling Z1 in transfected HEK 293 cells and native tissue. Taken together, we have established an integrated and generic purification strategy for the identification of high quality and functionally relevant GPCR-associated protein complexes that significantly widens the repertoire of available techniques.
Figures
Fig. 1.
Optimization of the peptide affinity chromatography conditions. A, determination of the optimal concentration of imidazole to minimize nonspecific binding on the beads. Different amounts of brain proteins were added to non-coated beads (20 μl) and incubated overnight at 4 °C in the presence or absence of 10 or 20 m
m
imidazole. After washes, proteins were eluted from the beads, and the nonspecific binding was quantified. B, determination of the optimal amount of brain protein lysates. Non-coated beads (20 μl) were incubated with 2, 4, 6, 8, or 10 mg of protein lysates in the presence of 20 m
m
imidazole overnight at 4 °C. After washes, proteins were eluted from the beads, and the nonspecific binding was quantified. C, determination of the binding kinetics of the His6 (6xHis)-tagged peptides to the beads. 500 μg of His6-tagged peptides were dissolved in a binding buffer containing 20 m
m
NaH2PO4, 6
m
urea, pH 8, at 1 mg/ml. Absorbance of the peptide solutions was measured at 280 nm after 15, 30, 60, and 90 min of incubation with beads. D, determination of the amount of brain proteins specifically recruited by the C-tails of MT1 and MT2. 10 mg of brain protein lysate were incubated with 20 μl of beads coated with His6-tagged baits in the presence of 20 m
m
imidazole overnight at 4 °C. After washes, the amount of retained proteins was quantified. C, non-coated beads. Results are expressed as mean ± S.E. (n = 5 for non-coated beads, n = 3 for MT1, and n = 6 for MT2).
Fig. 2.
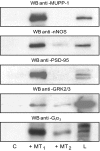
Detection of PDZ domain-containing proteins, Gi3α, and GRK2/3 by immunoblotting. 20 μl of beads coated with His6-tagged MT1 and MT2 C-tails were incubated with 10 mg of protein lysates from mouse brain. Beads were washed, and recruited proteins were eluted with 50 μl of 2% SDS in PBS. 10 μl were separated by SDS-PAGE and transferred to nitrocellulose membranes. Immunoblotting was performed with antibodies raised against three PDZ domain-containing proteins, polyclonal anti-MUPP1 (1:10,000), polyclonal anti-nNOS (1:1000) and monoclonal anti-PSD-95 (1:2000); against Gi3α (1:1000); and against GRK2/3 (1:1000). C, negative control, beads without peptide; L, brain lysate (20 μg). WB, Western blot.
Fig. 3.
2D and 1D electrophoresis separation of the MT1 and MT2 C-tail-associated protein complexes. Mouse brain protein complexes recruited by the Ni-NTA-immobilized C-tail of MT1 and MT2 receptors were separated by 2D (A) or 1D electrophoresis on a 10% (B) or 5–9% gradient polyacrylamide gel (C) and silver-stained. A typical gel for each condition is shown. C, negative control, beads without peptide. The arrows indicate the position of RGSZ1.
Fig. 4.
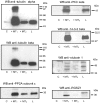
Validation of MALDI-TOF-identified binding proteins by immunoblotting. Immobilized His6-tagged MT1 and MT2 C-tails were incubated with 10 mg of protein lysates from mouse brain. Recruited proteins were eluted with 50 μl of 2% SDS in PBS. 10 μl were separated by SDS-PAGE and transferred to nitrocellulose membranes. Immunoblotting was performed with monoclonal anti-tubulin α (1:2000), monoclonal anti-tubulin β (1:2000), monoclonal anti-PP2A catalytic subunit (1:1000), polyclonal anti-PKC ζ (1:1000), monoclonal anti-14-3-3 β (1:1000), polyclonal anti-otubain 1 (1:2000), and polyclonal anti-murine RGSZ1 (1:500) antibodies. C, negative control, non-coated beads; L, brain lysate (20 μg). WB, Western blot.
Fig. 5.
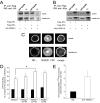
Functional interaction between RGSZ1 and MT1 in cells. HEK 293 cells transiently expressing either FLAG-MT1 or FLAG-MT2 were co-transfected with HA-RGSZ1 (A) or Myc-RGS10 (B). 48 h post-transfection, cells were stimulated, or not, by melatonin (10−7
m
for 15 min) and lysed. The MT1 and MT2 receptors were immunoprecipitated by anti-FLAG antibodies, and precipitates were analyzed by Western blot for the presence of co-immunoprecipitated HA-RGSZ1 (A) or Myc-RGS10 (B) using anti-tag antibodies. C, confocal images of HEK 293 cells co-expressing FLAG-MT1 with RGSZ1-YFP. Receptors were immunostained with anti-FLAG antibodies, and RGSZ1 was revealed by its YFP fluorescence. The co-localization of both proteins was evaluated with the ImageJ co-localization highlighter plug-in. D, [35S]GTPγS binding to MT1-expressing CHO cells without (black bars) or with 1 μ
m
melatonin (white bars) in the absence or presence of the indicated concentrations of purified RGSZ1. Data represent the mean ± S.E. of three experiments, each conducted in triplicate. Statistical significance was determined using the Mann-Whitney test. *, p < 0.05. E, co-immunoprecipitation of 125I-melatonin (125 I-MLT)-labeled MT1 receptors with anti-RGSZ1 antibodies. Nonspecific binding was estimated by performing immunoprecipitation using a pool of five non-relevant polyclonal antibodies. Data represent the mean ± S.E. of two experiments. IP, immunoprecipitate; WB, Western blot; NS, nonspecific binding.
Similar articles
- Purification and identification of G protein-coupled receptor protein complexes under native conditions.
Daulat AM, Maurice P, Froment C, Guillaume JL, Broussard C, Monsarrat B, Delagrange P, Jockers R. Daulat AM, et al. Mol Cell Proteomics. 2007 May;6(5):835-44. doi: 10.1074/mcp.M600298-MCP200. Epub 2007 Jan 9. Mol Cell Proteomics. 2007. PMID: 17215244 - Peptide affinity purification for the isolation and identification of GPCR-associated protein complexes.
Maurice P, Daulat AM, Jockers R. Maurice P, et al. Methods Mol Biol. 2011;746:389-98. doi: 10.1007/978-1-61779-126-0_22. Methods Mol Biol. 2011. PMID: 21607870 - Tandem affinity purification and identification of GPCR-associated protein complexes.
Daulat AM, Maurice P, Jockers R. Daulat AM, et al. Methods Mol Biol. 2011;746:399-409. doi: 10.1007/978-1-61779-126-0_23. Methods Mol Biol. 2011. PMID: 21607871 - Recent methodological advances in the discovery of GPCR-associated protein complexes.
Daulat AM, Maurice P, Jockers R. Daulat AM, et al. Trends Pharmacol Sci. 2009 Feb;30(2):72-8. doi: 10.1016/j.tips.2008.10.009. Epub 2008 Dec 25. Trends Pharmacol Sci. 2009. PMID: 19100631 Review. - Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry.
Völkel P, Le Faou P, Angrand PO. Völkel P, et al. Biochem Soc Trans. 2010 Aug;38(4):883-7. doi: 10.1042/BST0380883. Biochem Soc Trans. 2010. PMID: 20658971 Review.
Cited by
- Systematic protein-protein interaction mapping for clinically relevant human GPCRs.
Sokolina K, Kittanakom S, Snider J, Kotlyar M, Maurice P, Gandía J, Benleulmi-Chaachoua A, Tadagaki K, Oishi A, Wong V, Malty RH, Deineko V, Aoki H, Amin S, Yao Z, Morató X, Otasek D, Kobayashi H, Menendez J, Auerbach D, Angers S, Pržulj N, Bouvier M, Babu M, Ciruela F, Jockers R, Jurisica I, Stagljar I. Sokolina K, et al. Mol Syst Biol. 2017 Mar 15;13(3):918. doi: 10.15252/msb.20167430. Mol Syst Biol. 2017. PMID: 28298427 Free PMC article. - Melatonin and pancreatic islets: interrelationships between melatonin, insulin and glucagon.
Peschke E, Bähr I, Mühlbauer E. Peschke E, et al. Int J Mol Sci. 2013 Mar 27;14(4):6981-7015. doi: 10.3390/ijms14046981. Int J Mol Sci. 2013. PMID: 23535335 Free PMC article. Review. - Melatonin receptors: molecular pharmacology and signalling in the context of system bias.
Cecon E, Oishi A, Jockers R. Cecon E, et al. Br J Pharmacol. 2018 Aug;175(16):3263-3280. doi: 10.1111/bph.13950. Epub 2017 Aug 17. Br J Pharmacol. 2018. PMID: 28707298 Free PMC article. Review. - Update on melatonin receptors: IUPHAR Review 20.
Jockers R, Delagrange P, Dubocovich ML, Markus RP, Renault N, Tosini G, Cecon E, Zlotos DP. Jockers R, et al. Br J Pharmacol. 2016 Sep;173(18):2702-25. doi: 10.1111/bph.13536. Epub 2016 Aug 8. Br J Pharmacol. 2016. PMID: 27314810 Free PMC article. Review. - Identification of TSPAN4 as Novel Histamine H4 Receptor Interactor.
Ma X, Verweij EWE, Siderius M, Leurs R, Vischer HF. Ma X, et al. Biomolecules. 2021 Jul 30;11(8):1127. doi: 10.3390/biom11081127. Biomolecules. 2021. PMID: 34439793 Free PMC article.
References
- Fredriksson, R., Lagerstrom, M. C., Lundin, L. G., and Schioth, H. B. ( 2003) The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 - PubMed
- Vassilatis, D. K., Hohmann, J. G., Zeng, H., Li, F., Ranchalis, J. E., Mortrud, M. T., Brown, A., Rodriguez, S. S., Weller, J. R., Wright, A. C., Bergmann, J. E., and Gaitanaris, G. A. ( 2003) The G protein-coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. U. S. A. 100, 4903–4908 - PMC - PubMed
- Ellis, C., and The Nature Reviews Drug Discovery GPCR Questionnaire Participants ( 2004) The state of GPCR research in 2004. Nat. Rev. Drug Discov. 3, 575, 577–626 - PubMed
- Milligan, G., and White, J. H. ( 2001) Protein-protein interactions at G-protein-coupled receptors. Trends Pharmacol. Sci. 22, 513–518 - PubMed
- Dev, K. K., Nakanishi, S., and Henley, J. M. ( 2001) Regulation of mglu7 receptors by proteins that interact with the intracellular C-terminus. Trends Pharmacol. Sci. 22, 355–361 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous