Context-specific metabolic networks are consistent with experiments - PubMed (original) (raw)
Comparative Study
Context-specific metabolic networks are consistent with experiments
Scott A Becker et al. PLoS Comput Biol. 2008.
Abstract
Reconstructions of cellular metabolism are publicly available for a variety of different microorganisms and some mammalian genomes. To date, these reconstructions are "genome-scale" and strive to include all reactions implied by the genome annotation, as well as those with direct experimental evidence. Clearly, many of the reactions in a genome-scale reconstruction will not be active under particular conditions or in a particular cell type. Methods to tailor these comprehensive genome-scale reconstructions into context-specific networks will aid predictive in silico modeling for a particular situation. We present a method called Gene Inactivity Moderated by Metabolism and Expression (GIMME) to achieve this goal. The GIMME algorithm uses quantitative gene expression data and one or more presupposed metabolic objectives to produce the context-specific reconstruction that is most consistent with the available data. Furthermore, the algorithm provides a quantitative inconsistency score indicating how consistent a set of gene expression data is with a particular metabolic objective. We show that this algorithm produces results consistent with biological experiments and intuition for adaptive evolution of bacteria, rational design of metabolic engineering strains, and human skeletal muscle cells. This work represents progress towards producing constraint-based models of metabolism that are specific to the conditions where the expression profiling data is available.
Conflict of interest statement
BOP and UCSD have a financial interest in Genomatica, Inc. The findings in this manuscript may not benefit Genomatica, Inc.
Figures
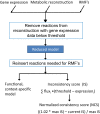
Figure 1. A flow chart schematic representation of the GIMME algorithm.
The GIMME algorithm takes three inputs: gene expression (or any other data type) mapped to reactions, a metabolic reconstruction, and one or more RMFs. A metabolic reconstruction is mapped through a data set, removing reactions that are not available and creating a reduced model. Reactions are reinserted into the reduced model as needed to achieve RMFs (such as growth and/or ATP production), resulting in a functional, context-specific model that features minimal disagreement with the data. The consistency score quantifies the disagreement with data, showing the minimal sum of fluxes weighted with reaction data deviations from data.
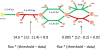
Figure 2. The computation of inconsistency scores.
Inconsistency scores for each reaction are computed by multiplying the deviation from a threshold by the required flux through a reaction. In the example here, the green reactions have data above the threshold, set to 12 (this is a parameter; see text). The red reactions have data below the threshold (11.4 and 8.2). The calculation of the inconsistency score corresponding to each reaction is shown numerically as flux multiplied by the deviation from the cutoff. They each increase the inconsistency score, implying that the data are less consistent with the objective of growing on lactate. Greater required fluxes and greater deviation from the threshold both increase the inconsistency scores. The total inconsistency score is the sum of all individual reaction scores.
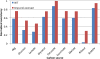
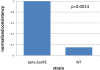
Figure 3. Glycerol-evolved strain normalized consistency scores.
Normalized consistency scores are computed directly from the inconsistency scores, as described in the text. A higher normalized consistency score indicates that the gene expression data is relatively more consistent with the RMF. Thus, here the gene expression data from the glycerol-evolved strains are more consistent with highly efficient growth on each of the carbon sources tested. The p values, determined by permutation testing, are less than 0.01 in all cases here.
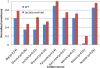
Figure 4. Lactate-evolved strain consistency scores.
This figure demonstrates the same result as Figure 3, but with strains evolved on lactate. The normalized consistency scores for growth on each of the tested carbon sources are higher for evolved strains, indicating that the gene expression data from the evolved strains are more consistent with efficient growth on each carbon source.
Figure 5. Metabolic engineering strain consistency score.
The normalized consistency score for an E. coli strain designed to produce lactate indicate that the Δpta ΔadhE strain has a metabolic gene expression state consistent with the simultaneous production of lactate and growth when compared with the wild-type. This higher normalized consistency score indicates that the gene expression data from the double deletion strain is more consistent with the metabolic engineering objective than the wild-type strain, in accordance with experimental measurements.
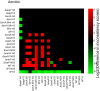
Figure 6. Pairwise comparisons of consistency for aerobic conditions.
A graphical representation of the log2 transform of the difference between inconsistency scores. A green box indicates that the sample on the _y_-axis is more consistent with aerobic growth than the sample on the _x_-axis. Red boxes indicate the opposite. Differences that do not meet p<0.05 are left blank. The shade of red or green quantifies the log2 of the difference in inconsistency scores. The position of green and red blocks here indicates that in all statistically significant cases, strains grown with oxygen have gene expression more consistent with efficient aerobic growth than strains grown without oxygen.
Figure 7. Pairwise comparisons of consistency for anaerobic conditions.
A graphical representation of the log2 transform of the difference between inconsistency scores. A green box indicates that the sample on the _y_-axis is more consistent with anaerobic growth than the sample on the _x_-axis. Red boxes indicate the opposite. Differences that do not meet p<0.05 are left blank. The shade of red or green quantifies the log2 of the difference in inconsistency scores. The position of green and red blocks shows that in nearly all cases that are statistically significant, gene expression data for strains grown without oxygen is more consistent with efficient anaerobic growth than strains grown with oxygen.
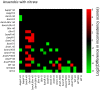
Figure 8. Pairwise comparisons of consistency for nitrate conditions.
A graphical representation of the log2 transform of the difference between inconsistency scores. A green box indicates that the sample on the _y_-axis is more consistent with nitrate growth than the sample on the _x_-axis. Red boxes indicate the opposite. Differences that do not meet p<0.05 are left blank. The shade of red or green quantifies the log2 of the difference in inconsistency scores. The position of green and red blocks indicates that in most cases, gene expression from strains grown with nitrate as the terminal electron acceptor is more consistent with efficient growth under this condition than strains grown under other conditions.
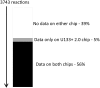
Figure 9. The mapping of Affymetrix gene chip data to reactions.
Reactions in the white area have no usable gene chip data on either platform. Reactions in grey have usable data only on the 133+ 2.0 platform. Reactions in black have usable data for both the 133+ 2.0 and the 133A platform. Importantly, 5% (179) of the reactions are only represented on the 133+ 2.0 chip, potentially increasing scores across chips. The average difference score is 340, so a difference of 179 reactions is greater than a 50% impact.
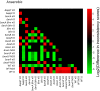
Figure 10. A comparison of skeletal muscle models.
This heat map displays the level of difference in each pair of models. Darker squares represent models that are more similar to each other than lighter squares. A black square (as on the diagonal) indicates identical models, and a white square indicates the most different pair of models. The three darker blocks that surround the main diagonal are the comparisons of samples within each dataset to each other. These darker blocks show that the models within each dataset tend to be more similar to each other than to models from other datasets. The models from a particular expression array type also appear to be more similar to each other than to models from a different array types, but the data available do not allow us to show that this is actually true, as is shown in Figure 11.
Figure 11. A comparison of skeletal muscle models, using only reactions that have data for both gene chips used.
This figure is the same as Figure 10, but the distances that are graphically represented are computed using only reactions that have data on both types of gene chips. The 5% of reactions that are represented on the U133+ 2.0 chip but not the U133A chip are not used for comparison. Here we no longer see any bias based on chip type, but rather we see that the FO datasets appear to be similar to both GI and GB data sets, instead of just the GI data sets. The chip type does not appear to affect the distances as much as the different experiments do. The chip type does appear to affect the reactions that the algorithm defines as active versus inactive, as seen in the differences between Figures 10 and 11.
Similar articles
- Computational methods for metabolic reconstruction.
Pitkänen E, Rousu J, Ukkonen E. Pitkänen E, et al. Curr Opin Biotechnol. 2010 Feb;21(1):70-7. doi: 10.1016/j.copbio.2010.01.010. Epub 2010 Feb 18. Curr Opin Biotechnol. 2010. PMID: 20171871 - Reconstruction of metabolic networks from high-throughput metabolite profiling data: in silico analysis of red blood cell metabolism.
Nemenman I, Escola GS, Hlavacek WS, Unkefer PJ, Unkefer CJ, Wall ME. Nemenman I, et al. Ann N Y Acad Sci. 2007 Dec;1115:102-15. doi: 10.1196/annals.1407.013. Epub 2007 Oct 9. Ann N Y Acad Sci. 2007. PMID: 17925356 - Data integration and analysis of biological networks.
Kim TY, Kim HU, Lee SY. Kim TY, et al. Curr Opin Biotechnol. 2010 Feb;21(1):78-84. doi: 10.1016/j.copbio.2010.01.003. Epub 2010 Feb 6. Curr Opin Biotechnol. 2010. PMID: 20138751 - Metabolic flux analysis in eukaryotes.
Niklas J, Schneider K, Heinzle E. Niklas J, et al. Curr Opin Biotechnol. 2010 Feb;21(1):63-9. doi: 10.1016/j.copbio.2010.01.011. Epub 2010 Feb 15. Curr Opin Biotechnol. 2010. PMID: 20163950 Review. - Inference of gene regulatory networks using boolean-network inference methods.
Hickman GJ, Hodgman TC. Hickman GJ, et al. J Bioinform Comput Biol. 2009 Dec;7(6):1013-29. doi: 10.1142/s0219720009004448. J Bioinform Comput Biol. 2009. PMID: 20014476 Review.
Cited by
- A network perspective on metabolic inconsistency.
Sonnenschein N, Golib Dzib JF, Lesne A, Eilebrecht S, Boulkroun S, Zennaro MC, Benecke A, Hütt MT. Sonnenschein N, et al. BMC Syst Biol. 2012 May 14;6:41. doi: 10.1186/1752-0509-6-41. BMC Syst Biol. 2012. PMID: 22583819 Free PMC article. - Contextualization procedure and modeling of monocyte specific TLR signaling.
Aurich MK, Thiele I. Aurich MK, et al. PLoS One. 2012;7(12):e49978. doi: 10.1371/journal.pone.0049978. Epub 2012 Dec 6. PLoS One. 2012. PMID: 23236359 Free PMC article. - scFASTCORMICS: A Contextualization Algorithm to Reconstruct Metabolic Multi-Cell Population Models from Single-Cell RNAseq Data.
Pacheco MP, Ji J, Prohaska T, García MM, Sauter T. Pacheco MP, et al. Metabolites. 2022 Dec 2;12(12):1211. doi: 10.3390/metabo12121211. Metabolites. 2022. PMID: 36557249 Free PMC article. - Integrative Gene Expression and Metabolic Analysis Tool IgemRNA.
Grausa K, Mozga I, Pleiko K, Pentjuss A. Grausa K, et al. Biomolecules. 2022 Apr 16;12(4):586. doi: 10.3390/biom12040586. Biomolecules. 2022. PMID: 35454176 Free PMC article. - How to understand the cell by breaking it: network analysis of gene perturbation screens.
Markowetz F. Markowetz F. PLoS Comput Biol. 2010 Feb 26;6(2):e1000655. doi: 10.1371/journal.pcbi.1000655. PLoS Comput Biol. 2010. PMID: 20195495 Free PMC article. No abstract available.
References
- Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nat Rev Genet. 2006;7:130–41. - PubMed
- Seshasayee AS, Bertone P, Fraser GM, Luscombe NM. Transcriptional regulatory networks in bacteria: From input signals to output responses. Curr Opin Microbiol. 2006;9:511–519. - PubMed
- Das D, Zhang MQ. Predictive models of gene regulation: Application of regression methods to microarray data. Methods Mol Biol. 2007;377:95–110. - PubMed
- Covert MW, Palsson BO. Transcriptional regulation in constraints-based metabolic models of Escherichia coli. J Biol Chem. 2002;277:28058–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources