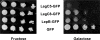Legionella eukaryotic-like type IV substrates interfere with organelle trafficking - PubMed (original) (raw)
Legionella eukaryotic-like type IV substrates interfere with organelle trafficking
Karim Suwwan de Felipe et al. PLoS Pathog. 2008.
Erratum in
- PLoS Pathog. 2010;6(3). doi: 10.1371/annotation/c7438c1b-5b65-4f4c-aaa3-062909e89525
- PLoS Pathog. 2010;6(3). doi: 10.1371/annotation/f079872b-7bce-41fe-bc16-c2e45d44b19e
Abstract
Legionella pneumophila, the causative agent of Legionnaires' disease, evades phago-lysosome fusion in mammalian and protozoan hosts to create a suitable niche for intracellular replication. To modulate vesicle trafficking pathways, L. pneumophila translocates effector proteins into eukaryotic cells through a Type IVB macro-molecular transport system called the Icm-Dot system. In this study, we employed a fluorescence-based translocation assay to show that 33 previously identified Legionella eukaryotic-like genes (leg) encode substrates of the Icm-Dot secretion system. To assess which of these proteins may contribute to the disruption of vesicle trafficking, we expressed each gene in yeast and looked for phenotypes related to vacuolar protein sorting. We found that LegC3-GFP and LegC7/YlfA-GFP caused the mis-secretion of CPY-Invertase, a fusion protein normally restricted to the yeast vacuole. We also found that LegC7/YlfA-GFP and its paralog LegC2/YlfB-GFP formed large structures around the yeast vacuole while LegC3-GFP localized to the plasma membrane and a fragmented vacuole. In mammalian cells, LegC2/YlfB-GFP and LegC7/YlfA-GFP were found within large structures that co-localized with anti-KDEL antibodies but excluded the lysosomal marker LAMP-1, similar to what is observed in Legionella-containing vacuoles. LegC3-GFP, in contrast, was observed as smaller structures which had no obvious co-localization with KDEL or LAMP-1. Finally, LegC3-GFP caused the accumulation of many endosome-like structures containing undigested material when expressed in the protozoan host Dictyostelium discoideum. Our results demonstrate that multiple Leg proteins are Icm/Dot-dependent substrates and that LegC3, LegC7/YlfA, and LegC2/YlfB may contribute to the intracellular trafficking of L. pneumophila by interfering with highly conserved pathways that modulate vesicle maturation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
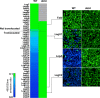
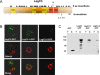
Figure 1. Icm-Dot Dependent Translocation Leg Substrates into J774 Cells.
J774 cells were infected with KS79 (WT) or KS79_dotA_ harboring TEM1-Leg protein fusions at an MOI of 50. Infected cells were loaded with CCF4/AM and translocation was determined by measuring the ratio of cleaved (460 nm) to uncleaved (530 nm) CCF4/AM. Ratios of 460/530 nm emission for each TEM1-Leg protein in wild type KS79 (WT) or KS79 dotA is shown as a heat diagram. A 460/530 nm ratio of more than 1 indicates translocation and is indicated by a horizontal dashed line. Representative images obtained using epifluorescence microscopy on individual assay wells is shown on the right. The results shown represent the average of 2 to 3 experiments, each performed in triplicate.
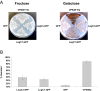
Figure 2. Effector-Induced Lethality in Yeast.
Yeast cells containing expression vectors for LegC5-GFP, LegC8-GFP, LepB-GFP fusion proteins or GFP alone were spotted in 10-fold serial dilutions on SC-ura/fructose or SC-ura/galactose plates. Under conditions that induce expression of the hybrid protein (galactose), LegC5-GFP, LegC8-GFP and LepB-GFP impair yeast growth. Under non-inducing conditions (fructose), all strains grow comparably well.
Figure 3. LegC3 and LegC7 Induce a Vacuolar Protein Sorting Defect in Yeast.
(A) Yeast strains expressing CPY-Inv and LegC3-GFP, LegC7-GFP, GFP or VPS4E233Q were streaked on SC-ura/fructose or SC-ura/galactose and grown for four days at 30°C. As described in the Materials and Methods, an agar solution containing chromogenic reagents to detect invertase activity was overlaid on the plates. The secretion of the CPY-Invertase hybrid is detected by the formation of a brown precipitate. (B) Quantitative assay for invertase secretion. Liquid cultures were grown to stationary phase and secreted versus total invertase activity was measured as described in Material and Methods. The results shown are of a representative experiment done in triplicate +/− standard deviations.
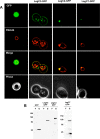
Figure 4. Steady State Localization of Leg-GFP Hybrids and Vacuole Defects.
(A) Yeast strains expressing the LegC2-GFP, LegC3-GFP, LegC7-GFP hybrid proteins and GFP alone were harvested and pulse-chased with N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl)-pyridinium dibromide (FM4-64) for vacuole visualization. Cells were viewed using epifluorescence microscopy. The images shown are representative of the overall population of cells expressing the Leg-GFP hybrids. (B) Whole cell lysate immunoblot using a rabbit polyclonal antibody against GFP showing that all the GFP Leg fusion proteins are expressed under galactose induction, but not when grown in fructose. The asterisk (*) is placed to emphasize that ten times more LegC7-GFP sample had to be loaded for visualization.
Figure 5. A Hydrophobic Region at the C-terminus of LegC3 is Essential for its Localization in Yeast.
(A) LegC3 was mutagenized using the GPS-LS Linker Scanning kit (New England Biolabs) to generate a library of 60 distinct mutant proteins with five amino acid insertions and 27 distinct truncation products. The positions of the 60 insertions are indicated by diamonds and the 27 truncations by triangles together with the LegC3 structural predictions. The open diamonds and triangles represent insertion and truncation sites, respectively, that exhibited an altered LegC3-GFP localization. (B) Epifluorescence microscopy showing specific localization patterns for LegC3-1 and LegC3-2 compared to wild-type LegC3. (C) Whole cell lysate immunoblot using a rabbit polyclonal antibody against GFP showing that the LegC3-1 and LegC3-2 mutant proteins are stably expressed under galactose induction.
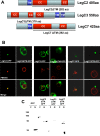
Figure 6. The Predicted Transmembrane Domains of LegC2, LegC3, and LegC7 are Required for Their Localization in Yeast.
(A) Constructs of LegC2, LegC3 and LegC7 were designed to exclude the predicted transmembrane regions. The solid line drawn beneath each gene represents the amino acid sequence included in each final construct. The truncated constructs were translationally fused to gfp and transformed into NSY01 for expression. (B) Epifluorescence microscopy showing localization patterns. (C) Whole cell lysate immunoblot using a rabbit polyclonal antibody against GFP showing the truncated proteins.
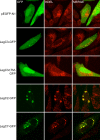
Figure 7. Localization of Leg-GFP Fusion Proteins and KDEL-containing proteins in transiently transfected CHO-FcγRII cells.
CHO-FcγRII cells were transfected with plasmids expressing GFP (pEGFP-N1), LegC3-GFP (pRG6), LegC3ΔTM (pRG7), LegC2-GFP (pRG8) or LegC7 (pRG10), as indicated on the left of each row. Representative confocal images were acquired demonstrating the location of GFP and proteins containing KDEL motifs as determined by immuno-fluorescent staining. Merged images are shown in the right column where yellow color designates overlap of the green and red channels.
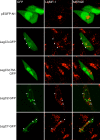
Figure 8. Localization of Leg-GFP Fusion Proteins and LAMP-1 in transiently transfected CHO-FcγRII cells.
CHO-FcγRII cells were transfected as in Figure 7. Representative confocal images demonstrating the location of GFP and LAMP-1 staining are shown. When present, arrows indicate the location of GFP-positive structures to distinguish these from proximal LAMP-1 lysosomes. Merged images are shown in the right column. The lack of yellow color in merged verifies a lack of colocalization between GFP-positive structures and lysosomes.
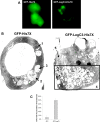
Figure 9. Expression of GFP-LegC3-His7X and GFP-His7X in D. discoideum.
(A) D. discoideum cells containing GFP-His7x, or GFP-LegC3-His7x under the control of a Tc-repressible promoter were grown with the presence of 10 µg/mL of Tc for 3 days at 25°C. The cells were washed in SorC buffer and incubated for an additional 8–10 hours in media without tetracycline. Expression of the fusion proteins was visualized under epifluorescence microscopy. (B) D. discoideum cells expressing hybrid fusion proteins were sorted for GFP+ cells after 10 hours of induction. The samples were then fixed and processed for electron microscopy using osmium tetroxide staining. Cells expressing GFP-His7X contain a mixture of vesicles with (1) undigested material, (2) partly digested material and (3) completely digested material. At least half of the cells expressing GFP-LegC3-His7X contain (4) an abnormally high number of vesicles with partly digested contents. What appears to be unfused pro-lysosomes (5) are segregated to a separate portion of the cell. (C) At least 100 cells of each sample were analyzed for abnormal fine structures. The percentage of GFP-LegC3-His7X expressing cells containing abnormal vesicle accumulation versus GFP-His7X expressing cells. 95% confidence intervals are indicated above.
Similar articles
- LegC3, an effector protein from Legionella pneumophila, inhibits homotypic yeast vacuole fusion in vivo and in vitro.
Bennett TL, Kraft SM, Reaves BJ, Mima J, O'Brien KM, Starai VJ. Bennett TL, et al. PLoS One. 2013;8(2):e56798. doi: 10.1371/journal.pone.0056798. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437241 Free PMC article. - The Legionella effector RavD binds phosphatidylinositol-3-phosphate and helps suppress endolysosomal maturation of the _Legionella_-containing vacuole.
Pike CM, Boyer-Andersen R, Kinch LN, Caplan JL, Neunuebel MR. Pike CM, et al. J Biol Chem. 2019 Apr 19;294(16):6405-6415. doi: 10.1074/jbc.RA118.007086. Epub 2019 Feb 7. J Biol Chem. 2019. PMID: 30733336 Free PMC article. - Legionella pneumophila Type IV Effectors YlfA and YlfB Are SNARE-Like Proteins that Form Homo- and Heteromeric Complexes and Enhance the Efficiency of Vacuole Remodeling.
Campodonico EM, Roy CR, Ninio S. Campodonico EM, et al. PLoS One. 2016 Jul 26;11(7):e0159698. doi: 10.1371/journal.pone.0159698. eCollection 2016. PLoS One. 2016. PMID: 27459495 Free PMC article. - Secreted phospholipases of the lung pathogen Legionella pneumophila.
Hiller M, Lang C, Michel W, Flieger A. Hiller M, et al. Int J Med Microbiol. 2018 Jan;308(1):168-175. doi: 10.1016/j.ijmm.2017.10.002. Epub 2017 Oct 28. Int J Med Microbiol. 2018. PMID: 29108710 Review. - Autophagy Evasion and Endoplasmic Reticulum Subversion: The Yin and Yang of Legionella Intracellular Infection.
Sherwood RK, Roy CR. Sherwood RK, et al. Annu Rev Microbiol. 2016 Sep 8;70:413-33. doi: 10.1146/annurev-micro-102215-095557. Annu Rev Microbiol. 2016. PMID: 27607556 Review.
Cited by
- Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Quaile AT, Urbanus ML, Stogios PJ, Nocek B, Skarina T, Ensminger AW, Savchenko A. Quaile AT, et al. Structure. 2015 Aug 4;23(8):1459-1469. doi: 10.1016/j.str.2015.05.020. Epub 2015 Jul 2. Structure. 2015. PMID: 26146184 Free PMC article. - Refining the plasmid-encoded type IV secretion system substrate repertoire of Coxiella burnetii.
Maturana P, Graham JG, Sharma UM, Voth DE. Maturana P, et al. J Bacteriol. 2013 Jul;195(14):3269-76. doi: 10.1128/JB.00180-13. Epub 2013 May 17. J Bacteriol. 2013. PMID: 23687269 Free PMC article. - A protein-protein interaction map reveals that the Coxiella burnetii effector CirB inhibits host proteasome activity.
Fu M, Liu Y, Wang G, Wang P, Zhang J, Chen C, Zhao M, Zhang S, Jiao J, Ouyang X, Yu Y, Wen B, He C, Wang J, Zhou D, Xiong X. Fu M, et al. PLoS Pathog. 2022 Jul 11;18(7):e1010660. doi: 10.1371/journal.ppat.1010660. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35816513 Free PMC article. - Genome-scale identification of Legionella pneumophila effectors using a machine learning approach.
Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. Burstein D, et al. PLoS Pathog. 2009 Jul;5(7):e1000508. doi: 10.1371/journal.ppat.1000508. Epub 2009 Jul 10. PLoS Pathog. 2009. PMID: 19593377 Free PMC article. - Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila.
Hervet E, Charpentier X, Vianney A, Lazzaroni JC, Gilbert C, Atlan D, Doublet P. Hervet E, et al. Infect Immun. 2011 May;79(5):1936-50. doi: 10.1128/IAI.00805-10. Epub 2011 Feb 14. Infect Immun. 2011. PMID: 21321072 Free PMC article.
References
- Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, et al. Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–1197. - PubMed
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, et al. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med. 1977;297:1197–1203. - PubMed
- Garcia-Vidal C, Carratala J. Current clinical management of Legionnaires' disease. Expert Rev Anti Infect Ther. 2006;4:995–1004. - PubMed
- Greenberg D, Chiou CC, Famigilleti R, Lee TC, Yu VL. Problem pathogens: paediatric legionellosis–implications for improved diagnosis. Lancet Infect Dis. 2006;6:529–535. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous