Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors - PubMed (original) (raw)
Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors
Veena Nukoolkarn et al. J Theor Biol. 2008.
Abstract
Since the emergence of the severe acute respiratory syndrome (SARS) to date, neither an effective antiviral drug nor a vaccine against SARS is available. However, it was found that a mixture of two HIV-1 proteinase inhibitors, lopinavir and ritonavir, exhibited some signs of effectiveness against the SARS virus. To understand the fine details of the molecular interactions between these proteinase inhibitors and the SARS virus via complexation, molecular dynamics simulations were carried out for the SARS-CoV 3CL(pro) free enzyme (free SARS) and its complexes with lopinavir (SARS-LPV) and ritonavir (SARS-RTV). The results show that flap closing was clearly observed when the inhibitors bind to the active site of SARS-CoV 3CL(pro). The binding affinities of LPV and RTV to SARS-CoV 3CL(pro) do not show any significant difference. In addition, six hydrogen bonds were detected in the SARS-LPV system, while seven hydrogen bonds were found in SARS-RTV complex.
Figures
Fig. 1
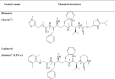
Chemical formula of HIV-1 PR inhibitor, ritonavir and lopinavir, which exhibits signs of effectives against SARS.
Fig. 2
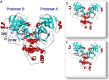
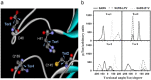
Structures of (a) SARS-free enzyme, (b) SARS–lopinavir complex and (c) SARS–ritronavir complex.
Fig. 3
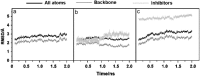
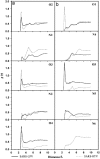
RMSDs of SARS-CoV 3CLpro free enzyme ((a) SARS) and its complexes with lopinavir ((b) SARS–LPV) and ritonavir ((c) SARS–RTV).
Fig. 4
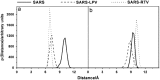
The probability distributions of the associated interatomic distances between the distances (a) _d_1 and (b) _d_2 from the center of mass of the following residues, H41 to C145 and D48 to Q189, respectively.
Fig. 5
(a) Definitions and (b) probability distributions of the torsion angles of the two catalytic residues (Tor1: CA-CB-CG-CD of H41 and TorR2: N-CA-CB-S of C145) and two amino acids at the tip of the loop (Tor3: CA-CB-CG-OD of D48 and Tor4: CB-CG-CD-CA of Q189) during 600–2000 ps of the simulations.
Fig. 6
Radial distribution functions, g(r), centered on the inhibitor atoms (see Fig. 1 for definitions) to oxygen atoms of modeled water of the two complexes, SARS–LPV and SARS–RTV. The plots are classified into two categories where the central atom types for the two inhibitors are topologically (a) equivalent and (b) different.
Similar articles
- Why Are Lopinavir and Ritonavir Effective against the Newly Emerged Coronavirus 2019? Atomistic Insights into the Inhibitory Mechanisms.
Nutho B, Mahalapbutr P, Hengphasatporn K, Pattaranggoon NC, Simanon N, Shigeta Y, Hannongbua S, Rungrotmongkol T. Nutho B, et al. Biochemistry. 2020 May 12;59(18):1769-1779. doi: 10.1021/acs.biochem.0c00160. Epub 2020 Apr 24. Biochemistry. 2020. PMID: 32293875 - Exploring the Binding Mechanism of PF-07321332 SARS-CoV-2 Protease Inhibitor through Molecular Dynamics and Binding Free Energy Simulations.
Ahmad B, Batool M, Ain QU, Kim MS, Choi S. Ahmad B, et al. Int J Mol Sci. 2021 Aug 24;22(17):9124. doi: 10.3390/ijms22179124. Int J Mol Sci. 2021. PMID: 34502033 Free PMC article. - Molecular Docking of Azithromycin, Ritonavir, Lopinavir, Oseltamivir, Ivermectin and Heparin Interacting with Coronavirus Disease 2019 Main and Severe Acute Respiratory Syndrome Coronavirus-2 3C-Like Proteases.
Arouche TDS, Martins AY, Ramalho TC, Júnior RNC, Costa FLP, Filho TSA, Neto AMJC. Arouche TDS, et al. J Nanosci Nanotechnol. 2021 Apr 1;21(4):2075-2089. doi: 10.1166/jnn.2021.19029. J Nanosci Nanotechnol. 2021. PMID: 33500022 - Lopinavir/ritonavir: a protease inhibitor for HIV-1 treatment.
Barragan P, Podzamczer D. Barragan P, et al. Expert Opin Pharmacother. 2008 Sep;9(13):2363-75. doi: 10.1517/14656566.9.13.2363. Expert Opin Pharmacother. 2008. PMID: 18710360 Review. - Lopinavir/ritonavir: Repurposing an old drug for HIV infection in COVID-19 treatment.
Magro P, Zanella I, Pescarolo M, Castelli F, Quiros-Roldan E. Magro P, et al. Biomed J. 2021 Mar;44(1):43-53. doi: 10.1016/j.bj.2020.11.005. Epub 2020 Nov 10. Biomed J. 2021. PMID: 33608241 Free PMC article. Review.
Cited by
- Identification of steroidal cardenolides from Calotropis procera as novel HIV-1 PR inhibitors: A molecular docking & molecular dynamics simulation study.
Swapna K, Srujana M, Mamidala E. Swapna K, et al. Indian J Med Res. 2024 Jul;160(1):78-86. doi: 10.25259/IJMR_2115_23. Indian J Med Res. 2024. PMID: 39382500 Free PMC article. - Exploring of N-phthalimide-linked 1,2,3-triazole analogues with promising -anti-SARS-CoV-2 activity: synthesis, biological screening, and molecular modelling studies.
Aljuhani A, Alsehli M, Seleem MA, Alraqa SY, Ahmed HEA, Rezki N, Aouad MR. Aljuhani A, et al. J Enzyme Inhib Med Chem. 2024 Dec;39(1):2351861. doi: 10.1080/14756366.2024.2351861. Epub 2024 Jun 7. J Enzyme Inhib Med Chem. 2024. PMID: 38847308 Free PMC article. - Adverse drug reactions associated with COVID-19 management.
Chavda V, Dodiya P, Apostolopoulos V. Chavda V, et al. Naunyn Schmiedebergs Arch Pharmacol. 2024 Oct;397(10):7353-7376. doi: 10.1007/s00210-024-03137-0. Epub 2024 May 14. Naunyn Schmiedebergs Arch Pharmacol. 2024. PMID: 38743117 Review. - Preparing for the next viral threat with broad-spectrum antivirals.
Karim M, Lo CW, Einav S. Karim M, et al. J Clin Invest. 2023 Jun 1;133(11):e170236. doi: 10.1172/JCI170236. J Clin Invest. 2023. PMID: 37259914 Free PMC article. Review. - Paxlovid (Nirmatrelvir/Ritonavir): A new approach to Covid-19 therapy?
Hashemian SMR, Sheida A, Taghizadieh M, Memar MY, Hamblin MR, Bannazadeh Baghi H, Sadri Nahand J, Asemi Z, Mirzaei H. Hashemian SMR, et al. Biomed Pharmacother. 2023 Jun;162:114367. doi: 10.1016/j.biopha.2023.114367. Epub 2023 Feb 6. Biomed Pharmacother. 2023. PMID: 37018987 Free PMC article. Review.
References
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main protease (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. - PubMed
- Carlson H.A. Protein flexibility and drug design: how to hit a moving target. Curr. Opin. Chem. Biol. 2002;6:447–452. - PubMed
- Carlson H.A., McCammon J.A. Accommodating protein flexibility in computational drug design. Mol. Pharmacol. 2000;57:213–218. - PubMed
- Case D.A., Pearlman J.W., Caldwell T.E., Cheatham J., Jr., Wang W.S., Ross C.L., Simmerling T.A., Darden K.M., Merz R.V., Stanton A.L., Cheng J.J., Vincent M., Crowley V., Tsui H., Gohlke R.J., Radmer Y., Duan J., Pitera I., Massova G.L., Seibel U.C., Singh P.K., Kollman P.A. University of California; San Francisco, CA: 2002. AMBER. (Version 7.0 ed.)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous