PRDM16 controls a brown fat/skeletal muscle switch - PubMed (original) (raw)
. 2008 Aug 21;454(7207):961-7.
doi: 10.1038/nature07182.
Bryan Bjork, Wenli Yang, Shingo Kajimura, Sherry Chin, Shihuan Kuang, Anthony Scimè, Srikripa Devarakonda, Heather M Conroe, Hediye Erdjument-Bromage, Paul Tempst, Michael A Rudnicki, David R Beier, Bruce M Spiegelman
Affiliations
- PMID: 18719582
- PMCID: PMC2583329
- DOI: 10.1038/nature07182
PRDM16 controls a brown fat/skeletal muscle switch
Patrick Seale et al. Nature. 2008.
Abstract
Brown fat can increase energy expenditure and protect against obesity through a specialized program of uncoupled respiration. Here we show by in vivo fate mapping that brown, but not white, fat cells arise from precursors that express Myf5, a gene previously thought to be expressed only in the myogenic lineage. We also demonstrate that the transcriptional regulator PRDM16 (PRD1-BF1-RIZ1 homologous domain containing 16) controls a bidirectional cell fate switch between skeletal myoblasts and brown fat cells. Loss of PRDM16 from brown fat precursors causes a loss of brown fat characteristics and promotes muscle differentiation. Conversely, ectopic expression of PRDM16 in myoblasts induces their differentiation into brown fat cells. PRDM16 stimulates brown adipogenesis by binding to PPAR-gamma (peroxisome-proliferator-activated receptor-gamma) and activating its transcriptional function. Finally, Prdm16-deficient brown fat displays an abnormal morphology, reduced thermogenic gene expression and elevated expression of muscle-specific genes. Taken together, these data indicate that PRDM16 specifies the brown fat lineage from a progenitor that expresses myoblast markers and is not involved in white adipogenesis.
Figures
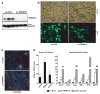
Fig. 1. Knockdown of PRDM16 in primary brown fat cells induces skeletal myogenesis
(a) Western blot analysis for PRDM16 in primary brown fat cell cultures transduced with adenovirus expressing shRNA targeted to PRDM16 or a scrambled (SCR) control shRNA. (b) These cultures were visualized by phase contrast microscopy and by GFP fluorescence. (c) Immunocytochemistry for skeletal Myosin Heavy Chain (MyHC) expression. (d) Gene expression at day 4 of adipocyte differentiation including BAT-selective and skeletal muscle-specific genes (as indicated). C2C12 myotubes were also assayed for their expression of muscle-specific genes (n=3, error bars represent ± SD; *p<0.05, **p<0.01).
Fig. 2. Brown fat and skeletal muscle arise from _myf5_-expressing precursors
(a)_myf5_-cre mice were intercrossed with indicator mice that have a YFP gene integrated into the rosa26 locus downstream of a floxed transcriptional stop sequence (R26R3-YFP). Expression of Cre recombinase excises the stop sequence to irreversibly activate YFP expression. (b) Immunohistochemistry to detect YFP (GFP) expression in skeletal muscle (sk. musc), BAT and WAT from control (_myf5_-cre negative) and myf5-cre:R26R3-YFP mice. (c) Real-time PCR analysis of YFP mRNA levels in: liver; gluteal (glut), inguinal (ing) and epididymal (epid) WAT; skeletal muscle (skM) and BAT (n= 4/group; error bars are ± SEM; **p<0.01). (d) UCP1 and GFP expression in WAT and BAT from CL316, 243 treated myf5-cre:R26R3-YFP mice.
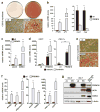
Fig. 3. PRDM16 stimulates brown adipocyte differentiation in skeletal myoblasts
(a–d) C2C12 myoblasts expressing retroviral PRDM16 or vector control (ctl) were stained with Oil-Red-O 6 days after inducing adipocyte differentiation (a); and analyzed by real-time PCR for their expression of markers specific to: adipocytes (PPARγ, aP2) (b, left); skeletal muscle (myod, myg) (b, right); BAT (elovl3, CIDEA) (c); and thermogenesis (UCP1, PGC-1α) (d). (e–g) ctl and PRDM16-expressing primary myoblasts were stained with Oil-Red-O 7 days after inducing adipocyte differentiation (e). Real-time PCR analysis of genes expressed selectively in adipocytes (PPARγ, aP2, adiponectin) (left); BAT (elovl3, CIDEA) and during thermogenesis (PGC-1α, UCP1). (g) Western blot analysis before (day 0) and after 7 days of differentiation. (n=4; error bars are ± SD; **p<0.05).
Fig. 4. PRDM16 binds and activates the transcriptional function of PPARγ
(a) Components in the PRDM16 complex from fat cells were separated by SDS-PAGE and visualized by silver staining. (b) Immunoprecipitation of PRDM16 from COS-7 cells expressing exogenous PRDM16 and/or Flag-PPARγ2 followed by western blot analysis to detect PPARγ2. (c) GST alone or a GST fusion protein containing PPARγ2 was incubated with 35S-labeled PRDM16 or SRC-1 protein (+/− 1 μM rosiglitazone). (d) GST fusion proteins containing different regions of PRDM16 were incubated with 35S-labeled PPARγ2. (e) Transcriptional activity of a PPAR-driven reporter gene in response to PPARγ/RXRα and PRDM16 or vector expression in COS-7 cells (+/− 1 μM rosiglitazone) (n=3; error bars are ± SD; **p<0.05).
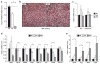
Fig. 5. Altered morphology and dysregulated gene expression in _PRDM16_-deficient brown fat
**(a)**Real-time PCR analysis of PRDM16 mRNA levels in putative BAT depots from E17 wildtype (Wt), heterozygous (het) and PRDM16 knock-out (KO) mice. (b) Hematoxylin and Eosin (H&E) staining of representative sections of BAT from Wt and KO mice. (c–e) Wt, het and KO BAT were examined by real-time PCR for their expression of: general adipocyte markers (aP2 and adiponectin) (c); BAT-selective genes (d); and skeletal muscle-specific genes (e). (n= 7–11 mice per group; error bars represent ± SEM). (*p < 0.05; **p < 0.01).
Comment in
- Developmental biology: Neither fat nor flesh.
Cannon B, Nedergaard J. Cannon B, et al. Nature. 2008 Aug 21;454(7207):947-8. doi: 10.1038/454947a. Nature. 2008. PMID: 18719573 No abstract available.
Similar articles
- Transcriptional control of brown fat determination by PRDM16.
Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Seale P, et al. Cell Metab. 2007 Jul;6(1):38-54. doi: 10.1016/j.cmet.2007.06.001. Cell Metab. 2007. PMID: 17618855 Free PMC article. - Peroxisome proliferator-activated receptor α (PPARα) induces PPARγ coactivator 1α (PGC-1α) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16.
Hondares E, Rosell M, Díaz-Delfín J, Olmos Y, Monsalve M, Iglesias R, Villarroya F, Giralt M. Hondares E, et al. J Biol Chem. 2011 Dec 16;286(50):43112-22. doi: 10.1074/jbc.M111.252775. Epub 2011 Oct 27. J Biol Chem. 2011. PMID: 22033933 Free PMC article. - PRDM16: the interconvertible adipo-myocyte switch.
Frühbeck G, Sesma P, Burrell MA. Frühbeck G, et al. Trends Cell Biol. 2009 Apr;19(4):141-6. doi: 10.1016/j.tcb.2009.01.007. Epub 2009 Mar 13. Trends Cell Biol. 2009. PMID: 19285866 - Role of PRDM16 in the activation of brown fat programming. Relevance to the development of obesity.
Becerril S, Gómez-Ambrosi J, Martín M, Moncada R, Sesma P, Burrell MA, Frühbeck G. Becerril S, et al. Histol Histopathol. 2013 Nov;28(11):1411-25. doi: 10.14670/HH-28.1411. Epub 2013 Jun 17. Histol Histopathol. 2013. PMID: 23771475 Review. - Brown vs white adipocytes: the PPARgamma coregulator story.
Koppen A, Kalkhoven E. Koppen A, et al. FEBS Lett. 2010 Aug 4;584(15):3250-9. doi: 10.1016/j.febslet.2010.06.035. Epub 2010 Jun 30. FEBS Lett. 2010. PMID: 20600006 Review.
Cited by
- MicroRNA-21 modulates brown adipose tissue adipogenesis and thermogenesis in a mouse model of polycystic ovary syndrome.
Rezq S, Huffman AM, Basnet J, Alsemeh AE, do Carmo JM, Yanes Cardozo LL, Romero DG. Rezq S, et al. Biol Sex Differ. 2024 Jul 10;15(1):53. doi: 10.1186/s13293-024-00630-2. Biol Sex Differ. 2024. PMID: 38987854 Free PMC article. - Selegiline Modulates Lipid Metabolism by Activating AMPK Pathways of Epididymal White Adipose Tissues in HFD-Fed Obese Mice.
Joung HY, Oh JM, Song MS, Kwon YB, Chun S. Joung HY, et al. Pharmaceutics. 2023 Oct 27;15(11):2539. doi: 10.3390/pharmaceutics15112539. Pharmaceutics. 2023. PMID: 38004519 Free PMC article. - Construction of a doxycycline inducible adipogenic lentiviral expression system.
Liu Q, Hill PJ, Karamitri A, Ryan KJ, Chen HY, Lomax MA. Liu Q, et al. Plasmid. 2013 Jan;69(1):96-103. doi: 10.1016/j.plasmid.2012.10.001. Epub 2012 Oct 23. Plasmid. 2013. PMID: 23099229 Free PMC article. - SMAD3 negatively regulates serum irisin and skeletal muscle FNDC5 and peroxisome proliferator-activated receptor γ coactivator 1-α (PGC-1α) during exercise.
Tiano JP, Springer DA, Rane SG. Tiano JP, et al. J Biol Chem. 2015 Mar 20;290(12):7671-84. doi: 10.1074/jbc.M114.617399. Epub 2015 Feb 3. J Biol Chem. 2015. PMID: 25648888 Free PMC article. - Programming human pluripotent stem cells into white and brown adipocytes.
Ahfeldt T, Schinzel RT, Lee YK, Hendrickson D, Kaplan A, Lum DH, Camahort R, Xia F, Shay J, Rhee EP, Clish CB, Deo RC, Shen T, Lau FH, Cowley A, Mowrer G, Al-Siddiqi H, Nahrendorf M, Musunuru K, Gerszten RE, Rinn JL, Cowan CA. Ahfeldt T, et al. Nat Cell Biol. 2012 Jan 15;14(2):209-19. doi: 10.1038/ncb2411. Nat Cell Biol. 2012. PMID: 22246346 Free PMC article.
References
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. - PubMed
- Cousin B, et al. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci. 1992;103(Pt 4):931–42. - PubMed
- Garruti G, Ricquier D. Analysis of uncoupling protein and its mRNA in adipose tissue deposits of adult humans. Int J Obes Relat Metab Disord. 1992;16:383–90. - PubMed
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR044031/AR/NIAMS NIH HHS/United States
- R01 AR044031-11/AR/NIAMS NIH HHS/United States
- R37 DK031405/DK/NIDDK NIH HHS/United States
- R37 DK031405-27/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases