Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo - PubMed (original) (raw)
Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo
Vesna Milacic et al. Cancer Res. 2008.
Abstract
Curcumin (diferuloylmethane) is the major active ingredient of turmeric (Curcuma longa) used in South Asian cuisine for centuries. Curcumin has been shown to inhibit the growth of transformed cells and to have a number of potential molecular targets. However, the essential molecular targets of curcumin under physiologic conditions have not been completely defined. Herein, we report that the tumor cellular proteasome is most likely an important target of curcumin. Nucleophilic susceptibility and in silico docking studies show that both carbonyl carbons of the curcumin molecule are highly susceptible to a nucleophilic attack by the hydroxyl group of the NH(2)-terminal threonine of the proteasomal chymotrypsin-like (CT-like) subunit. Consistently, curcumin potently inhibits the CT-like activity of a purified rabbit 20S proteasome (IC(50) = 1.85 micromol/L) and cellular 26S proteasome. Furthermore, inhibition of proteasome activity by curcumin in human colon cancer HCT-116 and SW480 cell lines leads to accumulation of ubiquitinated proteins and several proteasome target proteins, and subsequent induction of apoptosis. Furthermore, treatment of HCT-116 colon tumor-bearing ICR SCID mice with curcumin resulted in decreased tumor growth, associated with proteasome inhibition, proliferation suppression, and apoptosis induction in tumor tissues. Our study shows that proteasome inhibition could be one of the mechanisms for the chemopreventive and/or therapeutic roles of curcumin in human colon cancer. Based on its ability to inhibit the proteasome and induce apoptosis in both HCT-116 and metastatic SW480 colon cancer cell lines, our study suggests that curcumin could potentially be used for treatment of both early-stage and late-stage/refractory colon cancer.
Figures
Figure 1
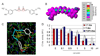
In silico and in vitro proteasome inhibition by curcumin. A, the chemical structure of curcumin. The regions with carbonyl carbons that have the SAR (structure activity relationship) are marked with red circles. B, molecular orbital energy analysis is demonstrated by drawing and electron density isosurface and coloring by nucleophilic susceptibility. The yellow center signifies the highest area of susceptibility. C, docking analysis of curcumin, which is represented by the stick structure and the colors are representative of atom type (carbon, gray; oxygen, red; hydrogen, white; methyl, light blue). The dotted line in yellow represents the distance, in angstroms, of each of the carbonyl carbons to Thr 1, indicative of potential nucleophilic attack, and to Ser 96, indicative of potential hydrogen bonding. D, in vitro analysis using a 20S proteasome indicates that curcumin inhibits chymotrypsin-like, trypsin-like and PGPH-like activities with IC50 values of 1.85±0.35, 6.23±0.22 and 3.68±0.19 µM, respectively. Ethanol was used as a control (E). Columns, mean of representative independent triplicate experiments; bars, SD.
Figure 2
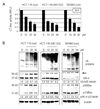
Inhibition of proteasomal chymotrypsin-like activity in human HCT-116 and SW480 colon cancer cells by curcumin. A, inhibition of proteasomal chymotrypsin-like activity by curcumin in intact HCT-116 and SW480 cells. HCT-116 and SW480 cells were treated with different curcumin concentrations (10, 20, and 30 µM) for 24 h, followed by measurement of proteasomal CT-like activity. Proteasome inhibitor MG132 was used as comparison with curcumin and the solvent, ethanol (E), was used as a control. B, Western blot analysis. HCT-116 and SW480 cells were treated with different curcumin concentrations (10, 20, and 30 µM) for 24 h, and used for whole cell extract preparation. Cell extract was analyzed by Western blot for accumulation of ubiquitinated proteins, IκB-α, and Bax (right and left panels). Proteasome inhibitor MG132 was used as comparison to curcumin (middle panel) and the solvent, ethanol (E), was used as a control. Actin was used as a loading control. Change in the levels of ubiquitinated IκB-α and p36/Bax proteins in the cells treated with curcumin, and the levels of ubiquitinated IκB-α and p21/Bax proteins in the cells treated with MG132 was analyzed by densitometry and quantified using AlphaEase FC software.
Figure 3
Induction of apoptosis in human HCT-116 and SW480 colon cancer cells by curcumin. A, A cell-free caspase-3/-7 activity assay. HCT-116 and SW480 cells were treated with different curcumin concentrations (10, 20, and 30 µM) for 24 h, harvested and prepared whole cell extract was used for fluorescent detection of caspase-3/-7 activity (see Materials and Methods). Proteasome inhibitor MG132 was used as comparison with curcumin and the solvent, ethanol (E), was used as a control. Columns, mean of representative independent triplicate experiments; bars, SD. B, Western blot analysis. Cell extract from HCT-116 and SW480 cells treated with curcumin was analyzed by Western blot for PARP cleavage and Cyclin D1 detection (left and right panels). Proteasome inhibitor MG132 was used as comparison with curcumin (middle panel) and the solvent, ethanol (E), was used as a control. Actin was used as a loading control. Change in Cyclin D1 protein level was analyzed by densitometry and quantified using AlphaEase FC software. C, For TUNEL assay, HCT-116 and SW480 cells were treated for 24 h with 30 µM curcumin or ethanol (solvent) as a control. The TUNEL assay was performed by using an APO-DIRECT kit and the number of apoptotic (TdT positive) cells is indicated.
Figure 4
Kinetic effect of curcumin on HCT-116 cells. A, HCT-116 cells were treated with 20 µM curcumin for 0.5 to 48 h, followed by the proteasomal chymotrypsin-like activity assay using Z-GGL-AMC. B, HCT-116 cells treated with 20 µM curcumin for 6 to 48 hours were used for whole cell extract preparation. Columns, mean of representative independent triplicate experiments; bars, SD. Cell extract analyzed by Western blot confirmed proteasomal inhibition by accumulation of ubiquitinated proteins, and proteasome target proteins IκB-α, p27, and p21/Bax. Apoptosis induced by curcumin treatment was confirmed by PARP cleavage (B), caspase-3/7 activation (C), and apoptotic morphological changes (D). Actin was used as a loading control. Columns, mean of independent triplicate experiments; bars, SD. Change in the level of IκB-α, p27, p21/Bax, p36/Bax and Cyclin D1 proteins was analyzed by densitometry and quantified using AlphaEase FC software.
Figure 5
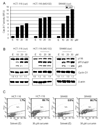
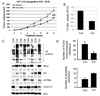
Curcumin treatment leads to inhibition of the proteasomal chymotrypsin-like activity, proliferation suppression and apoptosis induction in vivo. Female homozygous ICR SCID mice bearing HCT-116 tumors were treated with either control solvent or curcumin at 500 mg/kg/d to day 21. A, inhibition of HCT-116 tumor growth by curcumin. Points, mean tumor volume in each experimental group containing 6 mice; bars, SD; **, P <0.01. B–D, effects of curcumin at the end point of the experiment. Tumors were collected after 21 days of treatment, and the prepared tissues were analyzed by the proteasomal chymotrypsin-like activity assay (B), Western blotting (C), and immunohistochemistry (D). Inhibition of proteasome activity (B), accumulation of ubiquitinated proteins, p27, IκB-α, and p21/Bax proteins and down-regulation of Cyclin D1 (C) were found in all four tumors treated with curcumin (Cur), compared to control mice (Con) treated with the control solvent alone. The slides prepared from the tumors treated with the control solvent or curcumin were used for PCNA immunostaining and TUNEL (D). Bars represent mean numbers of PCNA immunoreactive cells per focus, SD (D, upper panel), and mean numbers of apoptotic cells per focus, SD (D, lower panel). Change in protein level was analyzed by densitometry and quantified using AlphaEase FC software.
Figure 6
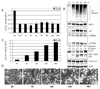
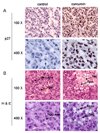
Immunohistochemistry p27 and Hematoxylin and Eosin (H & E) staining assays using mouse tumor samples. Tumors were collected after 21-day treatment (see Fig. 5 legend), and the prepared tissue slides were used for immunostaining with p27 antibody (A), and H & E staining assays (B). Stronger or/and more p27 positive cells (A), more apoptotic-condensed nuclei (Apo with arrow), and more necrotic tumor cells (Nec with arrow) were found in tumor tissue from mice treated with curcumin. Only few apoptotic cells and much more non-apoptotic cells (Non-apo with arrow) were found in tumor tissue from mice treated with solvent. Magnifications are 100X and 400X as indicated.
Similar articles
- Pristimerin induces apoptosis by targeting the proteasome in prostate cancer cells.
Yang H, Landis-Piwowar KR, Lu D, Yuan P, Li L, Reddy GP, Yuan X, Dou QP. Yang H, et al. J Cell Biochem. 2008 Jan 1;103(1):234-44. doi: 10.1002/jcb.21399. J Cell Biochem. 2008. PMID: 17541980 - Targeting proteasomal pathways by dietary curcumin for cancer prevention and treatment.
Hasima N, Aggarwal BB. Hasima N, et al. Curr Med Chem. 2014;21(14):1583-94. doi: 10.2174/09298673113206660135. Curr Med Chem. 2014. PMID: 23834173 Review. - Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2.
Banerjee S, Ji C, Mayfield JE, Goel A, Xiao J, Dixon JE, Guo X. Banerjee S, et al. Proc Natl Acad Sci U S A. 2018 Aug 7;115(32):8155-8160. doi: 10.1073/pnas.1806797115. Epub 2018 Jul 9. Proc Natl Acad Sci U S A. 2018. PMID: 29987021 Free PMC article. - The potential of proteasome inhibition in the treatment of colon cancer.
Konstantinopoulos PA, Papavassiliou AG. Konstantinopoulos PA, et al. Expert Opin Investig Drugs. 2006 Sep;15(9):1067-75. doi: 10.1517/13543784.15.9.1067. Expert Opin Investig Drugs. 2006. PMID: 16916273 Review.
Cited by
- The Efficacy and Safety of Highly-Bioavailable Curcumin for Treating Knee Osteoarthritis: A 6-Month Open-Labeled Prospective Study.
Nakagawa Y, Mukai S, Yamada S, Murata S, Yabumoto H, Maeda T, Akamatsu S. Nakagawa Y, et al. Clin Med Insights Arthritis Musculoskelet Disord. 2020 Aug 12;13:1179544120948471. doi: 10.1177/1179544120948471. eCollection 2020. Clin Med Insights Arthritis Musculoskelet Disord. 2020. PMID: 32848491 Free PMC article. - Curcumin ameliorates the neurodegenerative pathology in A53T α-synuclein cell model of Parkinson's disease through the downregulation of mTOR/p70S6K signaling and the recovery of macroautophagy.
Jiang TF, Zhang YJ, Zhou HY, Wang HM, Tian LP, Liu J, Ding JQ, Chen SD. Jiang TF, et al. J Neuroimmune Pharmacol. 2013 Mar;8(1):356-69. doi: 10.1007/s11481-012-9431-7. Epub 2013 Jan 17. J Neuroimmune Pharmacol. 2013. PMID: 23325107 - Biodegradable micelles enhance the antiglioma activity of curcumin in vitro and in vivo.
Zheng S, Gao X, Liu X, Yu T, Zheng T, Wang Y, You C. Zheng S, et al. Int J Nanomedicine. 2016 Jun 9;11:2721-36. doi: 10.2147/IJN.S102450. eCollection 2016. Int J Nanomedicine. 2016. PMID: 27354801 Free PMC article. - Biochemical and functional analyses of gp130 mutants unveil JAK1 as a novel therapeutic target in human inflammatory hepatocellular adenoma.
Poussin K, Pilati C, Couchy G, Calderaro J, Bioulac-Sage P, Bacq Y, Paradis V, Leteurtre E, Sturm N, Ramos J, Guettier C, Bardier-Dupas A, Boulai A, Wendum D, Selves J, Izard T, Nault JC, Zucman-Rossi J. Poussin K, et al. Oncoimmunology. 2013 Dec 1;2(12):e27090. doi: 10.4161/onci.27090. Epub 2014 Jan 3. Oncoimmunology. 2013. PMID: 24501689 Free PMC article. - The novel curcumin analog FLLL32 decreases STAT3 DNA binding activity and expression, and induces apoptosis in osteosarcoma cell lines.
Fossey SL, Bear MD, Lin J, Li C, Schwartz EB, Li PK, Fuchs JR, Fenger J, Kisseberth WC, London CA. Fossey SL, et al. BMC Cancer. 2011 Mar 28;11:112. doi: 10.1186/1471-2407-11-112. BMC Cancer. 2011. PMID: 21443800 Free PMC article.
References
- Somasundaram S, Edmund NA, Moore DT, et al. Dietary curcumin inhibits chemotherapy-induced apoptosis in models of human breast cancer. Cancer Res. 2002;62:3868–3875. - PubMed
- Goel A, Kunnumakkara AB, Aggarwal BB. Curcumin as “Curecumin”: from kitchen to clinic. Biochem Pharmacol. 2008;75:787–809. - PubMed
- Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776–4789. - PubMed
- Landis-Piwowar KR, Milacic V, Chen D, et al. The proteasome as a potential target for novel anticancer drugs and chemosensitizers. Drug Resist Updat. 2006;9:263–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA112625/CA/NCI NIH HHS/United States
- CA120009/CA/NCI NIH HHS/United States
- P30 CA022453/CA/NCI NIH HHS/United States
- R01 CA120009/CA/NCI NIH HHS/United States
- R03 CA112625/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources