The genetics of mammalian circadian order and disorder: implications for physiology and disease - PubMed (original) (raw)
Review
The genetics of mammalian circadian order and disorder: implications for physiology and disease
Joseph S Takahashi et al. Nat Rev Genet. 2008 Oct.
Abstract
Circadian cycles affect a variety of physiological processes, and disruptions of normal circadian biology therefore have the potential to influence a range of disease-related pathways. The genetic basis of circadian rhythms is well studied in model organisms and, more recently, studies of the genetic basis of circadian disorders has confirmed the conservation of key players in circadian biology from invertebrates to humans. In addition, important advances have been made in understanding how these molecules influence physiological functions in tissues throughout the body. Together, these studies set the scene for applying our knowledge of circadian biology to the understanding and treatment of a range of human diseases, including cancer and metabolic and behavioural disorders.
Figures
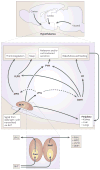
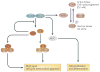
Figure 1. A schematic diagram of the SCN and its input and output pathways
The mammalian circadian pacemaker in the hypothalamic suprachiasmatic nucleus (SCN) is organized into two major subdivisions, the core and the shell. The core region of the SCN receives information about the daily light cycle via the retinohypothalamic tract (RHT). Both neuronal and humoral signals act as output signals from the SCN to other regions of the brain and the periphery. The SCN output pathways are responsible for proper timing of diverse physiological functions, including hormone release, sleep-wake cycle, feeding behavior, and thermoregulation. The SCN output to the subparaventricular zone (sPVz) is relayed to the medial preoptic region (MPO) to control circadian rhythms of body temperature; and a separate projection through the dorsomedial nucleus of the hypothalamus (DMH) controls daily hormone secretion (via paraventricular nucleus, PVN) and sleep-wake cycles (via lateral hypothalamus, LH, and ventrolateral preoptic nucleus, VLPO). Figure modified from [permission obtained].
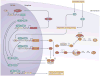
Figure 2. The mammalian circadian clock is composed of a transcriptional–translational feedback network
The circadian clock mechanism involves transcription-translation feedback loops comprised of a set of core clock genes. In mammals, the circadian clock is composed of a primary negative feedback loop involving the genes, Clock/Npas2, Bmal1, Period1 (Per1), Per2, Cryptochrome1 (Cry1) and Cry2. CLOCK/NPAS2 and BMAL1 are basic helix-loop-helix (bHLH) PAS-domain containing transcription factors that activate transcription of the Per and Cry genes. The resulting PER and CRY proteins heterodimerize, translocate to the nucleus and interact with the CLOCK:BMAL1 complex to inhibit their own transcription. With time, the PER-CRY repressor complex is degraded, and CLOCK:BMAL1 can then activate a new cycle of transcription. The secondary autoregulatory feedback loop is composed of REV-ERB_α_ that is a direct target of CLOCK:BMAL1 transcription activator complex. REV-ERB_α_ feeds back to repress Bmal1 transcription and competes with RORa to bind retinoic acid-related orphan receptor response elements (RREs) in the Bmal1 promoter. In addition to the transcriptional activators and repressors, post-translational modification and degradation of circadian clock proteins are critical steps for determining circadian periodicity. Key kinases for PER (and CRY) phosphorylation are Casein kinase 1δ (CK1δ) and CK1ε. One of the roles for phosphorylation of clock proteins is to target them for polyubiquitination and degradation by the 26S proteosomal pathway. Both β-TCRP1 and FBXL3 E3 ubiquitin ligase complexes have been implicated in targeting the PER and CRY proteins, respectively, for degradation. Figure modified from [permission sought].
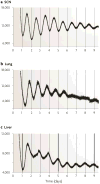
Figure 3. Central and peripheral oscillators
The expression of the core circadian clock genes is ubiquitous and reflects the presence of circadian oscillators in virtually every tissue and cell in the body. The SCN expresses robust oscillations of PER2::Luciferase activity when isolated in explant culture ex vivo, as do virtually every tissue in the mouse, such as the lung and the liver. Data from .
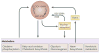
Figure 4. Interactions between circadian and metabolic systems
Fundamental metabolic pathways such as glycolysis, fatty acid metabolism, cholesterol biosynthesis, and xenobiotic and intermediate metabolism are all under circadian regulation. Metabolism itself can also have effects on circadian clocks: these include NADH and NADPH which can modulate CLOCK:BMAL1 and NPAS2:BMAL1 DNA binding; the PAS domains of NPAS2 which can bind heme and can be modulated by carbon monoxide; and, heme can also be a ligand for the circadian nuclear receptor, Rev-erbα.
Figure 5. Circadian control of cell division cycles
Circadian system is linked to the cell division cycle via circadian control of gene expression and posttranslational mechanisms. The transcription of c-Myc and Wee1 is circadian and appears to be a direct target of the CLOCK:BMAL1. The expression of Wee1 is co-regulated with Per and the entry of the cell cycle into M phase is suppressed during the day time when the transcription of Per (and Wee1) is high. In addition, the PER1 protein interacts with the check-point proteins, ATM and Chk2; while related work has linked the TIMELESS (TIM) and CRY proteins with Chk1. Activation of the DNA damage pathway can also reset the phase of the circadian clock.
Similar articles
- [Biology and genetics of circadian rhythm].
Bellivier F. Bellivier F. Encephale. 2009 Jan;35 Suppl 2:S53-7. doi: 10.1016/S0013-7006(09)75534-4. Encephale. 2009. PMID: 19268171 Review. French. - A molecular perspective of human circadian rhythm disorders.
Cermakian N, Boivin DB. Cermakian N, et al. Brain Res Brain Res Rev. 2003 Jun;42(3):204-20. doi: 10.1016/s0165-0173(03)00171-1. Brain Res Brain Res Rev. 2003. PMID: 12791440 Review. - [Synchronization and genetic redundancy in circadian clocks].
Dardente H. Dardente H. Med Sci (Paris). 2008 Mar;24(3):270-6. doi: 10.1051/medsci/2008243270. Med Sci (Paris). 2008. PMID: 18334175 Review. French. - [Timekeeping molecules: implications for circadian phenotypes].
Pereira DS, Tufik S, Pedrazzoli M. Pereira DS, et al. Braz J Psychiatry. 2009 Mar;31(1):63-71. doi: 10.1590/s1516-44462009000100015. Braz J Psychiatry. 2009. PMID: 19506779 Review. Portuguese. - Chronobiology in mammalian health.
Liu Z, Chu G. Liu Z, et al. Mol Biol Rep. 2013 Mar;40(3):2491-501. doi: 10.1007/s11033-012-2330-4. Epub 2012 Dec 6. Mol Biol Rep. 2013. PMID: 23224435 Review.
Cited by
- Synchronous activation of gonadotropin-releasing hormone gene transcription and secretion by pulsatile kisspeptin stimulation.
Choe HK, Kim HD, Park SH, Lee HW, Park JY, Seong JY, Lightman SL, Son GH, Kim K. Choe HK, et al. Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5677-82. doi: 10.1073/pnas.1213594110. Epub 2013 Mar 18. Proc Natl Acad Sci U S A. 2013. PMID: 23509283 Free PMC article. - Fractal patterns of neural activity exist within the suprachiasmatic nucleus and require extrinsic network interactions.
Hu K, Meijer JH, Shea SA, vanderLeest HT, Pittman-Polletta B, Houben T, van Oosterhout F, Deboer T, Scheer FA. Hu K, et al. PLoS One. 2012;7(11):e48927. doi: 10.1371/journal.pone.0048927. Epub 2012 Nov 20. PLoS One. 2012. PMID: 23185285 Free PMC article. - Circadian influences on dopamine circuits of the brain: regulation of striatal rhythms of clock gene expression and implications for psychopathology and disease.
Verwey M, Dhir S, Amir S. Verwey M, et al. F1000Res. 2016 Aug 24;5:F1000 Faculty Rev-2062. doi: 10.12688/f1000research.9180.1. eCollection 2016. F1000Res. 2016. PMID: 27635233 Free PMC article. Review. - The biological clock and the molecular basis of lysosomal storage diseases.
Mazzoccoli G, Mazza T, Vinciguerra M, Castellana S, Scarpa M. Mazzoccoli G, et al. JIMD Rep. 2015;18:93-105. doi: 10.1007/8904_2014_354. Epub 2015 Jan 13. JIMD Rep. 2015. PMID: 25583520 Free PMC article. - Electric light, particularly at night, disrupts human circadian rhythmicity: is that a problem?
Stevens RG, Zhu Y. Stevens RG, et al. Philos Trans R Soc Lond B Biol Sci. 2015 May 5;370(1667):20140120. doi: 10.1098/rstb.2014.0120. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 25780233 Free PMC article. Review.
References
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–61. - PubMed
- Antle MC, Silver R. Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 2005;28:145–51. - PubMed
- Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 MH078024/MH/NIMH NIH HHS/United States
- U01 MH61915/MH/NIMH NIH HHS/United States
- U01 MH061915/MH/NIMH NIH HHS/United States
- P50 MH074924/MH/NIMH NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources