The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes - PubMed (original) (raw)
The spindle assembly checkpoint is satisfied in the absence of interkinetochore tension during mitosis with unreplicated genomes
Christopher B O'Connell et al. J Cell Biol. 2008.
Abstract
The accuracy of chromosome segregation is enhanced by the spindle assembly checkpoint (SAC). The SAC is thought to monitor two distinct events: attachment of kinetochores to microtubules and the stretch of the centromere between the sister kinetochores that arises only when the chromosome becomes properly bioriented. We examined human cells undergoing mitosis with unreplicated genomes (MUG). Kinetochores in these cells are not paired, which implies that the centromere cannot be stretched; however, cells progress through mitosis. A SAC is present during MUG as cells arrest in response to nocodazole, taxol, or monastrol treatments. Mad2 is recruited to unattached MUG kinetochores and released upon their attachment. In contrast, BubR1 remains on attached kinetochores and exhibits a level of phosphorylation consistent with the inability of MUG spindles to establish normal levels of centromere tension. Thus, kinetochore attachment to microtubules is sufficient to satisfy the SAC even in the absence of interkinetochore tension.
Figures
Figure 1.
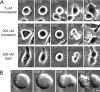
Spindle and kinetochore morphology of HeLa MUG. (A and B) Differential interference contrast (A) and centrin1-GFP (B) images of a fixed cell during MUG metaphase. (C–F) This cell was processed for correlative EM. (C) A single lower magnification section through the spindle. Note that chromatin is largely absent from the spindle except for a few small fragments (arrows). (D–F) 70-nm-thick serial sections through a kinetochore. Microtubules attach to the kinetochore end-on as well as laterally (E, arrow).
Figure 2.
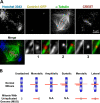
Geometries of kinetochore attachment in MUG. (A) Cells undergoing MUG were fixed and stained for chromatin, microtubules, and kinetochores. Images are maximum intensity projections of deconvolved z slices. Insets (single z slices) illustrate the types of microtubule–kinetochore attachments achievable in MUG: (1) merotelic, (2) monotelic, and (3) lateral. (B) Comparison of kinetochore attachments to spindle poles in normal mitosis versus MUG. Amphitelic or syntelic arrangements are not applicable (N.A.) for unpaired kinetochores.
Figure 3.
Behavior of unpaired kinetochores. (A) MUG in HeLa cells stably expressing CENP-A–GFP followed by combinational differential interference contrast (top) and three-dimensional fluorescence microscopy (bottom). Kinetochores are extremely dynamic, exhibiting rapid poleward and away from pole movements with a mean velocity of 1.8 ± 0.8 μm/min. Anaphase in this cell (56 s) is marked by cessation of kinetochore dynamics, spindle shortening (compare 49 s with 56 s), and outward movement of chromatin (arrowheads). Not all kinetochores congress before anaphase (arrow). (B) Tracking of a single kinetochore (arrowheads) in the cell shown in A. This as well as many other kinetochores exhibits rapid back and forth movement toward and away from the spindle equator. Time is given in hours/minutes. Bar, 2.5 μm.
Figure 4.
Evidence of a robust SAC during MUG. (A) HeLa cells undergoing MUG were treated with the indicated drugs and monitored by time-lapse microscopy. The disappearance of the nucleolus (arrowheads) indicates entry into MUG at t = 0. In the presence of nocodazole, there is a prolonged checkpoint arrest followed by escape that is marked by formation of micronuclei and respreading. Exit from MUG is also delayed when cells are treated with monastrol or taxol. Bar, 10 μm. (B) A cell in MUG after washout (t = 0) of 1.5 μM nocodazole. A bipolar spindle forms, and kinetochores congress to the equator (36 s). Soon after, the cell exits MUG, demonstrating satisfaction of the SAC. Time is given in hours/minutes.
Figure 5.
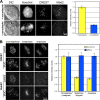
Behavior of Mad2 and BubR1 during MUG. (A) Cells in MUG were treated with nocodazole to generate unattached kinetochores or were left untreated. Differential interference contrast and fluorescence images of cells stained for DNA, kinetochores, and Mad2. The histogram presents the integrated intensity of Mad2 normalized to nocodazole-treated cells. (B) The distribution of BubR1 during normal mitosis and MUG. The histogram presents the integrated intensity of BubR1 on control and MUG kinetochores normalized to prometaphase. Error bars indicate SEM.
Similar articles
- Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores.
Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Hoffman DB, et al. Mol Biol Cell. 2001 Jul;12(7):1995-2009. doi: 10.1091/mbc.12.7.1995. Mol Biol Cell. 2001. PMID: 11451998 Free PMC article. - The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint.
Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. Hauf S, et al. J Cell Biol. 2003 Apr 28;161(2):281-94. doi: 10.1083/jcb.200208092. Epub 2003 Apr 21. J Cell Biol. 2003. PMID: 12707311 Free PMC article. - Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores.
Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Ditchfield C, et al. J Cell Biol. 2003 Apr 28;161(2):267-80. doi: 10.1083/jcb.200208091. J Cell Biol. 2003. PMID: 12719470 Free PMC article. - Tension sensors reveal how the kinetochore shares its load.
Salmon ED, Bloom K. Salmon ED, et al. Bioessays. 2017 Jul;39(7):10.1002/bies.201600216. doi: 10.1002/bies.201600216. Epub 2017 Jun 5. Bioessays. 2017. PMID: 28582586 Free PMC article. Review. - Microtubule-interfering agents, spindle defects, and interkinetochore tension.
Qi F, Zhou J, Liu M. Qi F, et al. J Cell Physiol. 2020 Jan;235(1):26-30. doi: 10.1002/jcp.28978. Epub 2019 Jun 20. J Cell Physiol. 2020. PMID: 31219174 Review.
Cited by
- Chromosome tips damaged in anaphase inhibit cytokinesis.
Baker NM, Zeitlin SG, Shi LZ, Shah J, Berns MW. Baker NM, et al. PLoS One. 2010 Aug 25;5(8):e12398. doi: 10.1371/journal.pone.0012398. PLoS One. 2010. PMID: 20811641 Free PMC article. - A Gradient in Metaphase Tension Leads to a Scaled Cellular Response in Mitosis.
Mukherjee S, Sandri BJ, Tank D, McClellan M, Harasymiw LA, Yang Q, Parker LL, Gardner MK. Mukherjee S, et al. Dev Cell. 2019 Apr 8;49(1):63-76.e10. doi: 10.1016/j.devcel.2019.01.018. Epub 2019 Feb 21. Dev Cell. 2019. PMID: 30799228 Free PMC article. - Mitotic spindle assembly around RCC1-coated beads in Xenopus egg extracts.
Halpin D, Kalab P, Wang J, Weis K, Heald R. Halpin D, et al. PLoS Biol. 2011 Dec;9(12):e1001225. doi: 10.1371/journal.pbio.1001225. Epub 2011 Dec 27. PLoS Biol. 2011. PMID: 22215983 Free PMC article. - Mitosis in vertebrates: the G2/M and M/A transitions and their associated checkpoints.
Rieder CL. Rieder CL. Chromosome Res. 2011 Apr;19(3):291-306. doi: 10.1007/s10577-010-9178-z. Chromosome Res. 2011. PMID: 21194009 Review. - Chromosomal instability: Stretching the role of checkpoint silencing.
Cheeseman LP, Maiato H. Cheeseman LP, et al. Curr Biol. 2021 Apr 26;31(8):R386-R389. doi: 10.1016/j.cub.2021.02.014. Curr Biol. 2021. PMID: 33905696 Free PMC article.
References
- Balczon, R.C. 2001. Overexpression of cyclin A in human HeLa cells induces detachment of kinetochores and spindle pole/centrosome overproduction. Chromosoma. 110:381–392. - PubMed
- Brinkley, B.R., R.P. Zinkowski, W.L. Mollon, F.M. Davis, M.A. Pisegna, M. Pershouse, and P.N. Rao. 1988. Movement and segregation of kinetochores experimentally detached from mammalian chromosomes. Nature. 336:251–254. - PubMed
- Ganem, N.J., K. Upton, and D.A. Compton. 2005. Efficient mitosis in human cells lacking polewards microtubule flux. Curr. Biol. 15:1827–1832. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM059363/GM/NIGMS NIH HHS/United States
- GM 59363/GM/NIGMS NIH HHS/United States
- F32 GM077911/GM/NIGMS NIH HHS/United States
- GM077911/GM/NIGMS NIH HHS/United States
- R01 GM059363-10/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases