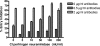Identification of major histocompatibility complex class I C molecule as an attachment factor that facilitates coronavirus HKU1 spike-mediated infection - PubMed (original) (raw)
Identification of major histocompatibility complex class I C molecule as an attachment factor that facilitates coronavirus HKU1 spike-mediated infection
Che Man Chan et al. J Virol. 2009 Jan.
Abstract
Human coronavirus HKU1 (HCoV-HKU1) is a recently discovered human coronavirus associated with respiratory tract infections worldwide. In this study, we have identified the major histocompatibility complex class I C molecule (HLA-C) as an attachment factor in facilitating HCoV-HKU1 spike (S)-mediated infection. HCoV-HKU1 S pseudotyped virus was assembled using a human immunodeficiency virus type 1-derived reporter virus harboring the human codon-optimized spike of HCoV-HKU1. We identified human alveolar epithelial A549 cells as the most susceptible cell line among those tested to infection by HCoV-HKU1 S pseudotypes. A549 cells were shown to bind purified soluble HCoV-HKU1 S(1-600) glycopeptide. To search for the functional receptor for HCoV-HKU1, an A549 cDNA expression library was constructed and transduced into the nonpermissive, baby hamster kidney cells line BHK-21. Transduced cells that bind soluble HCoV-HKU1 S(1-600) glycoprotein with C-terminal FLAG were sorted. Sequencing of two independent clones revealed cDNA inserts encoding HLA-C. Inhibition of HLA-C expression or function by RNAi silencing and anti-HLA-C antibody decreased HCoV-HKU1 S pseudotyped virus infection of A549 cells by 62 to 65%, whereas pretreatment of cells with neuraminidase decreased such infection by only 13%. When HLA-C was constitutively expressed in another nonpermissive cell line, NIH-3T3, quantitative PCR showed that the binding of HCoV-HKU1 S pseudotyped virus to cell surfaces was increased by 200-fold, but the cells remained nonsusceptible to HCoV-HKU1 S pseudotyped virus infection. Our data suggest that HLA-C is involved in the attachment of HCoV-HKU1 to A549 cells and is a potential candidate to facilitate cell entry. However, other unknown surface proteins on A549 cells may be concomitantly utilized by S glycoprotein of HCoV-HKU1 during viral entry. Further studies are required to elucidate other putative receptors or coreceptors for HCoV-HKU1 and the mechanism of HCoV-HKU1 S-mediated cell entry.
Figures
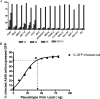
FIG. 1.
Tissue tropism demonstrated by infectivity of HCoV-HKU1 S-bearing pseudotyped virus in different cell lines. (A) Three different doses of pseudotyped viruses infected different cell lines at a cell density of 1 × 105 per well in a 24-well plate. Infectivity was measured by expression of the reporter eGFP by flow cytometry. VSV-G was included as a positive control. A total of 1× HKU1 pseudotyped virus is equivalent to 12.5 ng, quantified by detection of p24. Percentage of infection is measured by GFP expression of infected cells over the total cell population. (B) Infectivity of A549 cells by CoV-HKU1 S pseudotyped virus was viral load dependent, and saturation was achieved at ∼40 ng. The ACE2-transduced 293T cell line is a kind gift from M. Farzan (22).
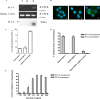
FIG. 2.
Expression of secreted S1 which can bind to A549 cells. (A) Secreted S1-600 was expressed as an 88-kDa N-linked glycoprotein which is Endo H resistant. +, test performed; −, test not performed. (B) S1-bound A549 cells were measured by flow cytometry using M2 anti-FLAG-FITC against C-terminally tagged FLAG (FACSCalibur; Becton Dickinson). Binding is inhibited by preincubated S1-600 with convalescent serum from a CoV-HKU1-infected patient (C), but not by normal-donor serum (D). (E) S1-600 does not bind to nonpermissive BHK-21 cells. Black line with gray-shaded area, A549 cell line; black line with white area, BHK-21 cell line; green line, S1 binding.
FIG. 3.
In vitro interaction of CoV-HKU1 S1-FLAG with HLA-C by coimmunoprecipitation. Purified S1-600-FLAG preadsorbed onto anti-FLAG M2 affinity beads was incubated with HLA-C-transfected BHK-21 cell lysate. Precipitant complexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed with anti-human HLA-C (lanes 1 to 3) and anti-FLAG-M2-HRP (lanes 4 to 6) by Western blotting (WB). BAP-FLAG (Sigma) and untransfected BHK-21 cell lysate were used as control. HLA-C-transfected BHK-21 cell lysate was recognized, pulled down by S1-FLAG, and coprecipitated and detected by both antibodies (lanes 1 and 4). No cross-reaction was observed between HLA-C and BAP-FLAG, and only BAP-FLAG was detected (lanes 2 and 5), while S1-FLAG did not show nonspecific recognition with untransfected cell lysate (lanes 3 and 6). +, components added; −, components not added.
FIG. 4.
Inhibition of HCoV-HKU1 S-pseudotyped virus entry into A549 cells by knockdown of HLA-C expression by siRNAs or by anti-HLA-C antibody. (A) A549 cells were transfected with siRNA1 (lane 1), siRNA2 (lane 2), and siRNA negative control (lane 3) and harvested for RT-PCR after 24 h. (B and C) A549 cells treated by stealth RNAi-1 (lane 1), stealth RNAi-2 against HLA-C (lane 2), and RNAi negative control (lane 3) for 24 h were fixed and surface stained by goat anti-human HLA-C (200 μg/ml; Santa Cruz) (immunofluorescence [IF], 1:100 and 1:150 for flow cytometry) and rabbit anti-goat IgG (H+L) FITC (IF, 1:500 and 1:100 for flow cytometry) and analyzed by immunofluorescent microscopy (Eclipse 80i Nikon) (B) and flow cytometry (FACSCalibur; Becton Dickinson) (C). (D and E) Effects on entry of CoV-HKU1 S pseudotyped virus. The nucleus was stained by DAPI in blue. A549 cells were either transfected with two individual duplexes of siRNAs for 24 h (D) or preincubated with polyclonal HLA-C antibodies at various concentrations as indicated for 1 h at 37°C (E). A total of 40 ng of CoV-HKU1 pseudotyped virus was inoculated onto 1 × 105 treated or untreated A549 cells in 24-well plates and incubated for 1 h at 37°C after three washes and being replenished with fresh medium. Percentage of infection was indicated by GFP expression and determined by flow cytometry. VSV-G pseudotypes were included as positive control.
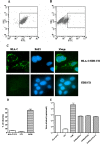
FIG. 5.
Expression of HLA-C does not restore CoV-HKU1 infection in nonpermissive cell lines but enhances viral binding to cell surfaces. HLA-C was stably expressed in HCoV-HKU1 nonpermissive cells. NIH-3T3 (3T3) cells were infected with VSV-G pseudotyped virus carrying human HLA-C/pFB-Neo and selected under Geneticin. NIH-3T3 cell lines constitutively expressing HLA-C were surface stained with anti-human HLA-C polyclonal antibodies and determined by flow cytometry (HLA-C/3T3 [A] and NIH-3T3 cells [B], respectively) and immunofluorescence (C). A total of 40 ng CoV-HKU1 pseudotyped virus was inoculated onto cells at the concentration of 1 × 105 cells/well in 24-well plates. For infectivity studies, virus was incubated with cells at 37°C for 1 h and washed and replenished with fresh medium. (D) GFP expression was measured by flow cytometry 48 h postinfection. For quantitation of viral attachment, pseudotyped virus was inoculated onto cells at 4°C for 1 h. After three washes, total RNA was harvested from cell lysis. (E) The number of viral copies was determined by real-time quantitative PCR.
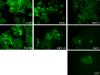
FIG. 6.
Immunofluorescence microscopy showing surface staining of HLA-C in tested human cell lines. All tested human cell lines were able to be stained positively for the presence of HLA-C. The differential intensities corroborate well with the relative MEFL ratios shown in Table 1.
FIG. 7.
Effect of neuraminidase treatment of A549 cells on CoV-HKU1 S pseudotyped virus infection. A549 cells grown in 24-well plates were incubated with the indicated concentrations of neuraminidase from C. perfringens (Cl perfringen) for 1 h prior to inoculation with 40 ng CoV-HKU1 S pseudotyped virus, with or without 1 h of preincubation with anti-HLA-C polyclonal antibody.
Similar articles
- Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme.
Huang X, Dong W, Milewska A, Golda A, Qi Y, Zhu QK, Marasco WA, Baric RS, Sims AC, Pyrc K, Li W, Sui J. Huang X, et al. J Virol. 2015 Jul;89(14):7202-13. doi: 10.1128/JVI.00854-15. Epub 2015 Apr 29. J Virol. 2015. PMID: 25926653 Free PMC article. - Identification of the Receptor-Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1.
Qian Z, Ou X, Góes LG, Osborne C, Castano A, Holmes KV, Dominguez SR. Qian Z, et al. J Virol. 2015 Sep;89(17):8816-27. doi: 10.1128/JVI.03737-14. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085157 Free PMC article. - Attachment glycoproteins and receptor specificity of rat coronaviruses.
Gagneten S, Scanga CA, Dveksler GS, Beauchemin N, Percy D, Holmes KV. Gagneten S, et al. Lab Anim Sci. 1996 Apr;46(2):159-66. Lab Anim Sci. 1996. PMID: 8723231 - Towards a coronavirus-based HIV multigene vaccine.
Eriksson KK, Makia D, Maier R, Ludewig B, Thiel V. Eriksson KK, et al. Clin Dev Immunol. 2006 Jun-Dec;13(2-4):353-60. doi: 10.1080/17402520600579168. Clin Dev Immunol. 2006. PMID: 17162377 Free PMC article. Review. - Identification of new human coronaviruses.
Pyrc K, Berkhout B, van der Hoek L. Pyrc K, et al. Expert Rev Anti Infect Ther. 2007 Apr;5(2):245-53. doi: 10.1586/14787210.5.2.245. Expert Rev Anti Infect Ther. 2007. PMID: 17402839 Review.
Cited by
- Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells.
Chu H, Chan CM, Zhang X, Wang Y, Yuan S, Zhou J, Au-Yeung RK, Sze KH, Yang D, Shuai H, Hou Y, Li C, Zhao X, Poon VK, Leung SP, Yeung ML, Yan J, Lu G, Jin DY, Gao GF, Chan JF, Yuen KY. Chu H, et al. J Biol Chem. 2018 Jul 27;293(30):11709-11726. doi: 10.1074/jbc.RA118.001897. Epub 2018 Jun 10. J Biol Chem. 2018. PMID: 29887526 Free PMC article. - Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1.
Ou X, Guan H, Qin B, Mu Z, Wojdyla JA, Wang M, Dominguez SR, Qian Z, Cui S. Ou X, et al. Nat Commun. 2017 May 23;8:15216. doi: 10.1038/ncomms15216. Nat Commun. 2017. PMID: 28534504 Free PMC article. - Emergence of Bat-Related Betacoronaviruses: Hazard and Risks.
Frutos R, Serra-Cobo J, Pinault L, Lopez Roig M, Devaux CA. Frutos R, et al. Front Microbiol. 2021 Mar 15;12:591535. doi: 10.3389/fmicb.2021.591535. eCollection 2021. Front Microbiol. 2021. PMID: 33790874 Free PMC article. Review. - Carcinoembryonic Antigen-Related Cell Adhesion Molecule 5 Is an Important Surface Attachment Factor That Facilitates Entry of Middle East Respiratory Syndrome Coronavirus.
Chan CM, Chu H, Wang Y, Wong BH, Zhao X, Zhou J, Yang D, Leung SP, Chan JF, Yeung ML, Yan J, Lu G, Gao GF, Yuen KY. Chan CM, et al. J Virol. 2016 Sep 29;90(20):9114-27. doi: 10.1128/JVI.01133-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27489282 Free PMC article. - Impact of HLA polymorphisms on the susceptibility to SARS-CoV-2 infection and related mortality in patients with renal replacement therapy.
Akcay OF, Yeter HH, Unsal Y, Yasar E, Gonen S, Derici U. Akcay OF, et al. Hum Immunol. 2023 Apr;84(4):272-277. doi: 10.1016/j.humimm.2023.01.008. Epub 2023 Feb 6. Hum Immunol. 2023. PMID: 36797091 Free PMC article.
References
- Chan, C. M., P. C. Woo, S. K. Lau, H. Tse, H. L. Chen, F. Li, B. J. Zheng, L. Chen, J. D. Huang, and K. Y. Yuen. Spike protein of human coronavirus HKU1: role in viral life cycle and antibody detection. Exp. Biol. Med. (Maywood), in press. - PubMed
- Chang, L. J., V. Urlacher, T. Iwakuma, Y. Cui, and J. Zucali. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 6715-728. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials