Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists - PubMed (original) (raw)
Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists
Da Wei Huang et al. Nucleic Acids Res. 2009 Jan.
Abstract
Functional analysis of large gene lists, derived in most cases from emerging high-throughput genomic, proteomic and bioinformatics scanning approaches, is still a challenging and daunting task. The gene-annotation enrichment analysis is a promising high-throughput strategy that increases the likelihood for investigators to identify biological processes most pertinent to their study. Approximately 68 bioinformatics enrichment tools that are currently available in the community are collected in this survey. Tools are uniquely categorized into three major classes, according to their underlying enrichment algorithms. The comprehensive collections, unique tool classifications and associated questions/issues will provide a more comprehensive and up-to-date view regarding the advantages, pitfalls and recent trends in a simpler tool-class level rather than by a tool-by-tool approach. Thus, the survey will help tool designers/developers and experienced end users understand the underlying algorithms and pertinent details of particular tool categories/tools, enabling them to make the best choices for their particular research interests.
Figures
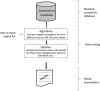
Figure 1.
The infrastructure of typical enrichment tools. Even though the enrichment analysis tools have distinct features, they can be generally described as three major layers: backend annotation database; data mining; and result presentation. Each of the layers, rather than statistical methods alone, greatly influences the analytic results.
Similar articles
- Extracting biological meaning from large gene lists with DAVID.
Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Huang da W, et al. Curr Protoc Bioinformatics. 2009 Sep;Chapter 13:Unit 13.11. doi: 10.1002/0471250953.bi1311s27. Curr Protoc Bioinformatics. 2009. PMID: 19728287 - GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists.
Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. Eden E, et al. BMC Bioinformatics. 2009 Feb 3;10:48. doi: 10.1186/1471-2105-10-48. BMC Bioinformatics. 2009. PMID: 19192299 Free PMC article. - GO FEAT: a rapid web-based functional annotation tool for genomic and transcriptomic data.
Araujo FA, Barh D, Silva A, Guimarães L, Ramos RTJ. Araujo FA, et al. Sci Rep. 2018 Jan 29;8(1):1794. doi: 10.1038/s41598-018-20211-9. Sci Rep. 2018. PMID: 29379090 Free PMC article. - How to learn about gene function: text-mining or ontologies?
Soldatos TG, Perdigão N, Brown NP, Sabir KS, O'Donoghue SI. Soldatos TG, et al. Methods. 2015 Mar;74:3-15. doi: 10.1016/j.ymeth.2014.07.004. Epub 2014 Aug 1. Methods. 2015. PMID: 25088781 Review. - Computational Methods and Software Tools for Functional Analysis of miRNA Data.
Garcia-Moreno A, Carmona-Saez P. Garcia-Moreno A, et al. Biomolecules. 2020 Aug 28;10(9):1252. doi: 10.3390/biom10091252. Biomolecules. 2020. PMID: 32872205 Free PMC article. Review.
Cited by
- Genome-Wide Association Study Reveals the Genetic Architecture of Growth and Meat Production Traits in a Chicken F2 Resource Population.
Volkova NA, Romanov MN, Vetokh AN, Larionova PV, Volkova LA, Abdelmanova AS, Sermyagin AA, Griffin DK, Zinovieva NA. Volkova NA, et al. Genes (Basel). 2024 Sep 25;15(10):1246. doi: 10.3390/genes15101246. Genes (Basel). 2024. PMID: 39457370 Free PMC article. - The TUTase URT1 connects decapping activators and prevents the accumulation of excessively deadenylated mRNAs to avoid siRNA biogenesis.
Scheer H, de Almeida C, Ferrier E, Simonnot Q, Poirier L, Pflieger D, Sement FM, Koechler S, Piermaria C, Krawczyk P, Mroczek S, Chicher J, Kuhn L, Dziembowski A, Hammann P, Zuber H, Gagliardi D. Scheer H, et al. Nat Commun. 2021 Feb 26;12(1):1298. doi: 10.1038/s41467-021-21382-2. Nat Commun. 2021. PMID: 33637717 Free PMC article. - Neurokinin-1 receptor signaling induces a pro-inflammatory transcriptomic profile in CD16+ monocytes.
Pappa V, Spitsin S, Gaskill PJ, Douglas SD. Pappa V, et al. J Neuroimmunol. 2021 Apr 15;353:577524. doi: 10.1016/j.jneuroim.2021.577524. Epub 2021 Feb 20. J Neuroimmunol. 2021. PMID: 33640716 Free PMC article. - Cytochrome P450 CYP78A9 is involved in Arabidopsis reproductive development.
Sotelo-Silveira M, Cucinotta M, Chauvin AL, Chávez Montes RA, Colombo L, Marsch-Martínez N, de Folter S. Sotelo-Silveira M, et al. Plant Physiol. 2013 Jun;162(2):779-99. doi: 10.1104/pp.113.218214. Epub 2013 Apr 22. Plant Physiol. 2013. PMID: 23610218 Free PMC article. - Germline copy number variation of genes involved in chromatin remodelling in families suggestive of Li-Fraumeni syndrome with brain tumours.
Aury-Landas J, Bougeard G, Castel H, Hernandez-Vargas H, Drouet A, Latouche JB, Schouft MT, Férec C, Leroux D, Lasset C, Coupier I, Caron O, Herceg Z, Frebourg T, Flaman JM. Aury-Landas J, et al. Eur J Hum Genet. 2013 Dec;21(12):1369-76. doi: 10.1038/ejhg.2013.68. Epub 2013 Apr 24. Eur J Hum Genet. 2013. PMID: 23612572 Free PMC article.
References
- Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. - PubMed
- Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. - PubMed
- Castillo-Davis CI, Hartl DL. GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical