Genome-wide loss of heterozygosity and uniparental disomy in BRCA1/2-associated ovarian carcinomas - PubMed (original) (raw)
Genome-wide loss of heterozygosity and uniparental disomy in BRCA1/2-associated ovarian carcinomas
Christine S Walsh et al. Clin Cancer Res. 2008.
Abstract
Purpose: The importance of the BRCA gene products in maintaining genomic stability led us to hypothesize that BRCA-associated and sporadic ovarian cancers would have distinctive genetic profiles despite similarities in histologic appearance.
Experimental design: A whole-genome copy number analysis of fresh, frozen, papillary serous ovarian cancer DNA was done using the Affymetrix 50K Xba Mapping Array using each patient's normal genomic DNA as the matched control. Loss of heterozygosity and copy number abnormalities were summarized to define regions of amplification, deletion, or uniparental disomy (UPD), defined as loss of one allele and duplication of the remaining allele. Genomic abnormalities were compared between BRCA-associated and sporadic tumors.
Results: We compared 6 BRCA-associated with 14 sporadic papillary serous ovarian carcinomas. Genetic instability, measured by percentage of genome altered, was more pronounced in BRCA-associated tumors (median, 86.6%; range, 54-100%) than sporadic tumors (median, 43.6%; range, 2-83%; P = 0.009). We used frequency plots to show the proportion of cases affected by each type abnormality at each genomic region. BRCA-associated tumors showed genome-wide loss of heterozygosity primarily due to the occurrence of UPD rather than deletion. UPD was found in 100% of the BRCA-associated and 50% of the sporadic tumors profiled.
Conclusions: This study reports on a previously underappreciated genetic phenomenon of UPD, which occurs frequently in ovarian cancer DNA. We observed distinct genetic patterns between BRCA-associated and sporadic ovarian cancers, suggesting that these papillary serous tumors arise from different molecular pathways.
Figures
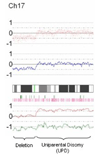
Fig. 1
Representative CNAG output. Elements from top to bottom include SNP plot (each red dot represents a SNP, baseline n=2), copy-number plot (blue line represents average copy number over consecutive SNPs, baseline n=2), chromosomal cytoband, heterozygosity calls (each green vertical bar represents a heterozygous, i.e. A/B, SNP call), loss of heterozygosity calls (each pink vertical bar represents LOH in tumor DNA compared to normal, i.e. A/A or B/B in tumor DNA; A/B in normal DNA), and allele-specific plot (red and green lines each represent specific allele copy number, baseline n=1). This example on chromosome 17 demonstrates deletion of the p-arm (copy number <2, one allele deleted, LOH) and UPD of the q-arm (copy number 2, one allele deleted, one allele duplicated, LOH).
Fig. 2
Box and whisker plot demonstrates significantly higher degree of genetic instability among BRCA-associated cases compared to sporadic cases, p=0.009. Median is represented by the white line, 25th and 75th percentiles (interquartile range, IQR) by the grey box, data range by the whiskers, and outliers (defined as more than 1.5IQR beyond the first or third quartile) are plotted as individual data points.
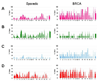
Fig. 3
Frequency plot summarizing the specific types of genetic changes occurring in sporadic versus BRCA-associated tumors: LOH (A, pink bars), deletions (B, green bars), uiparental disomy (C, blue bars), amplifications (D, red bars). In each panel, the percentage of cases affected by a specific type of genomic abnormality at each region is represented as a vertical bar along the y-axis. The genomic location is represented along the x-axis with chromosome 1 to the left and the X chromosome on the right.
Fig. 4
Genomic abnormalities on chromosomes 13 and 17 are demonstrated at higher resolution. Cases 1–6 correspond to the tumors from BRCA mutation carriers (see Table 1 for mutation details) while cases 7–20 represent the sporadic tumors. The types of abnormalities characterized include amplification (red), hemizygous deletion (green), UPD with diploid copy number (light blue), and UPD with copy number > 2 (dark blue). The locations of important tumor suppressor genes are shown with black hatch marks on the cytoband on the left edge of the graphs: BRCA2 (chromosome 13 at 31.7 MB), RB1 (chromosome 13 at 47.7 MB), TP53 (chromosome 17 at 7.5 MB) and BRCA1 (chromosome 17 at 38.5 MB).
Similar articles
- Gene expression profiles of BRCA1-linked, BRCA2-linked, and sporadic ovarian cancers.
Jazaeri AA, Yee CJ, Sotiriou C, Brantley KR, Boyd J, Liu ET. Jazaeri AA, et al. J Natl Cancer Inst. 2002 Jul 3;94(13):990-1000. doi: 10.1093/jnci/94.13.990. J Natl Cancer Inst. 2002. PMID: 12096084 - Gross genomic alterations and gene expression profiles of high- grade serous carcinoma of the ovary with and without BRCA1 inactivation.
Pradhan M, Risberg BA, Tropé CG, van de Rijn M, Gilks CB, Lee CH. Pradhan M, et al. BMC Cancer. 2010 Sep 15;10:493. doi: 10.1186/1471-2407-10-493. BMC Cancer. 2010. PMID: 20843305 Free PMC article. - Loss of heterozygosity on chromosome 13q12-q14, BRCA-2 mutations and lack of BRCA-2 promoter hypermethylation in sporadic epithelial ovarian tumors.
Gras E, Cortes J, Diez O, Alonso C, Matias-Guiu X, Baiget M, Prat J. Gras E, et al. Cancer. 2001 Aug 15;92(4):787-95. doi: 10.1002/1097-0142(20010815)92:4<787::aid-cncr1384>3.0.co;2-4. Cancer. 2001. PMID: 11550149 - The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer.
Lapunzina P, Monk D. Lapunzina P, et al. Biol Cell. 2011 Jul;103(7):303-17. doi: 10.1042/BC20110013. Biol Cell. 2011. PMID: 21651501 Review. - BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer.
Moschetta M, George A, Kaye SB, Banerjee S. Moschetta M, et al. Ann Oncol. 2016 Aug;27(8):1449-55. doi: 10.1093/annonc/mdw142. Epub 2016 Mar 31. Ann Oncol. 2016. PMID: 27037296 Review.
Cited by
- Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells.
Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, Chari R, Shames DS, Tang X, MacAulay C, Varella-Garcia M, Vooder T, Wistuba II, Lam S, Brekken R, Toyooka S, Minna JD, Lam WL, Gazdar AF. Soh J, et al. PLoS One. 2009 Oct 14;4(10):e7464. doi: 10.1371/journal.pone.0007464. PLoS One. 2009. PMID: 19826477 Free PMC article. - Relationship of microsatellite instability to mismatch repair deficiency in malignant tumors of dogs.
Inanaga S, Igase M, Sakai Y, Hagimori K, Sunahara H, Horikirizono H, Itamoto K, Baba K, Ohsato Y, Mizuno T. Inanaga S, et al. J Vet Intern Med. 2022 Sep;36(5):1760-1769. doi: 10.1111/jvim.16454. Epub 2022 Aug 11. J Vet Intern Med. 2022. PMID: 35959511 Free PMC article. - Acquired uniparental disomy of chromosome 9p in hematologic malignancies.
Wang L, Wheeler DA, Prchal JT. Wang L, et al. Exp Hematol. 2016 Aug;44(8):644-52. doi: 10.1016/j.exphem.2015.11.005. Epub 2015 Dec 2. Exp Hematol. 2016. PMID: 26646991 Free PMC article. Review. - Cross-laboratory validation of the OncoScan® FFPE Assay, a multiplex tool for whole genome tumour profiling.
Foster JM, Oumie A, Togneri FS, Vasques FR, Hau D, Taylor M, Tinkler-Hundal E, Southward K, Medlow P, McGreeghan-Crosby K, Halfpenny I, McMullan DJ, Quirke P, Keating KE, Griffiths M, Spink KG, Brew F. Foster JM, et al. BMC Med Genomics. 2015 Feb 18;8:5. doi: 10.1186/s12920-015-0079-z. BMC Med Genomics. 2015. PMID: 25889064 Free PMC article. - Mutant allele specific imbalance in oncogenes with copy number alterations: Occurrence, mechanisms, and potential clinical implications.
Yu CC, Qiu W, Juang CS, Mansukhani MM, Halmos B, Su GH. Yu CC, et al. Cancer Lett. 2017 Jan 1;384:86-93. doi: 10.1016/j.canlet.2016.10.013. Epub 2016 Oct 8. Cancer Lett. 2017. PMID: 27725226 Free PMC article. Review.
References
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. - PubMed
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. - PubMed
- Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. - PubMed
- Hoglund M, Gisselsson D, Hansen GB, Sall T, Mitelman F. Ovarian carcinoma develops through multiple modes of chromosomal evolution. Cancer Res. 2003;63:3378–3385. - PubMed
- Taetle R, Aickin M, Yang JM, et al. Chromosome abnormalities in ovarian adenocarcinoma: I. Nonrandom chromosome abnormalities from 244 cases. Genes Chromosomes Cancer. 1999;25:290–300. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous