Ribosome association and stability of the nascent polypeptide-associated complex is dependent upon its own ubiquitination - PubMed (original) (raw)
Ribosome association and stability of the nascent polypeptide-associated complex is dependent upon its own ubiquitination
Olesya O Panasenko et al. Genetics. 2009 Feb.
Erratum in
- Editor's Note: Ribosome Association and Stability of the Nascent Polypeptide-Associated Complex Is Dependent Upon Its Own Ubiquitination.
[No authors listed] [No authors listed] Genetics. 2025 Jan 8;229(1):1. doi: 10.1093/genetics/iyae177. Genetics. 2025. PMID: 39560658 No abstract available.
Abstract
In this work we addressed the role of ubiquitination in the function of the nascent polypeptide-associated complex (NAC), named EGD in the yeast Saccharomyces cerevisiae. To this end, we first identified the lysines residues required for ubiquitination of EGD/NAC. While simultaneous mutation of many lysines in the alpha-subunit of NAC (Egd2p) was required to abolish its ubiquitination, for the beta-subunit of NAC (Egd1p), mutation of K29 and K30 was sufficient. We determined that the ubiquitination of the two EGD subunits was coordinated, occurring during growth first on Egd1p and then on Egd2p. Egd2p was ubiquitinated earlier during growth if Egd1p could not be ubiquitinated. The use of mutants revealed the importance of EGD ubiqutination for its ribosome association and stability. Finally, our study demonstrated an interaction of EGD/NAC with the proteasome and revealed the importance of the Not4p E3 ligase, responsible for EGD/NAC ubiquitination, in this association.
Figures
Figure 1.—
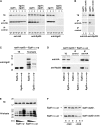
Different roles of Ubc4p and Ubc5p in Egd1p ubiquitination. (A) Total protein extracts were prepared from wild-type (WT), _ubc4_Δ, _ubc5_Δ, _not4_Δ, _ubc4_Δ _not4_Δ, and _ubc5_Δ _not4_Δ cells collected from exponential phase to glucose depletion and expressing Egd1p-HA3 and His6-ubiquitin. TEs and proteins eluted from a nickel column (Ni-eluate) were analyzed by Western blot with antibodies against the epitope tag. The molecular weights of protein markers are indicated on the left. (B) _ubc4_Δ or _ubc5_Δ strains expressing Myc6-Ubc4p or Myc6-Ubc5p were harvested after the indicated times in medium with cycloheximide (CHX, 50 μg/ml). The levels of the tagged proteins (indicated on the left) were revealed by rapid alkaline lysis and Western blot using antibodies against the tag (left), and equal protein loading was verified by ponceau S staining (right). (C) Total protein extracts prepared from _ubc4_Δ or _ubc5_Δ cells expressing Myc6-Ubc4p or Myc6-Ubc5p and His6-ubiquitin were collected from exponential phase to glucose depletion and analyzed as described in A.
Figure 2.—
Ubiquitination of Egd1p and Egd2p is linked. (A) Primary structure of Egd1p and Egd2p. The most conserved lysine residues are marked in boldface. NAC and UBA domains are underlined with one or two lines, respectively. The mapped ribosome-binding site in the structure of Egd1p is indicated by italics. (B) Total protein extracts were prepared from _egd1_Δ cells collected from exponential phase to glucose depletion and expressing from episomes His6-ubiquitin and HA-tagged Egd1p, either wild type or carrying the indicated mutations, and analyzed as in Figure 1A. Membranes were analyzed with antibodies against HA (top) or, after stripping, with antibodies against Egd2p (bottom). Molecular weights of protein markers are indicated on the left. (C) Total protein extracts were prepared from _egd2_Δ cells expressing from episomes His6-ubiquitin and HA-tagged Egd2p, either wild type or carrying the indicated mutations, and analyzed as described in Figure 1A. The signal in C for Egd2p is stronger than in B because different antibodies were used (HA vs. Egd2p). (D) Total protein extracts were prepared from _egd1_Δ (lanes 1 and 2) or _egd1_Δ _not4_Δ (lanes 3) cells expressing His6-ubiquitin and HA-tagged Egd1p, either wild type (lanes 1 and 3) or K29,30R (lane 2). Cells were grown and collected as described in Figure 1A. Molecular weights of protein markers are indicated on the left.
Figure 3.—
Stability of Egd1p is dependent upon its own ubiquitination and Egd2p. (A) _egd2_Δ cells expressing HA-tagged Egd2p, either wild type or carrying the indicated mutations, were treated and analyzed as in Figure 1B, with antibodies against HA or against Egd1p to reveal endogenous Egd1p. (B) _egd1_Δ cells expressing HA-tagged Egd1p, either wild type or carrying the indicated mutations, were treated and analyzed as in Figure 1B, with antibodies against HA or against Egd2p to reveal endogenous Egd2p. (C) _egd1_Δ _egd2_Δ cells expressing HA-tagged Egd1p, either wild type or carrying the indicated mutations, were analyzed as in Figure 1B. (D) _egd1_Δ _egd2_Δ cells expressing GFP-Egd2p or GFP-Egd2Δ_UBA_p and HA-tagged Egd1p, either wild type (WT), K10,13,29,30R (4R), or K10,13,29,30,43,50,66R (7R) were grown to an OD600 of 0.8 and collected after 6 hr when the drop of Egd1p in cells totally lacking Egd2p was maximal (C). Total extracts were analyzed by Western blot for the presence of the Egd1p and Egd2p derivatives with antibodies against the HA or GFP tags.
Figure 4.—
Ribosome association of the EGD complex is dependent upon its own ubiquitination and Egd2p. (A) Egd1p derivatives were immunoprecipitated from total protein extracts of _egd1_Δ or _egd1_Δ _egd2_Δ cells expressing tagged Egd1p, either wild type (WT, lanes 1), K10,13,29,30R (4R, lane 2, or K10,13,29,30,43,50,66R (7R, lane 3), with antibodies against the HA tag. TE and immunoprecipitates were analyzed by Western blot for the presence of tagged Egd1p (left), Rpl25p (middle), and Egd2p (right) with the antibodies indicated below the panels. (B) Egd1p-HA was immunoprecipitated from _egd1_Δ (lane 1) or _egd1_Δ _not4_Δ (lane 2) cells. TE and immunoprecipitates were analyzed for the presence of Egd1p-HA (left) or Rpl25p (right). (C) Total protein extracts were prepared from _egd1_Δ _egd2_Δ cells expressing His6-ubiquitin, Egd1K29,30Rp-HA, and wild-type or mutant Egd2p-HA as indicated. TE and proteins eluted from a nickel column were analyzed by Western blot with antibodies against Egd2p. The molecular weights of protein markers are indicated on the left. (D) The EGD complex was immunoprecipitated from total protein extracts collected from _egd1_Δ _egd2_Δ cells expressing Egd1K29,30Rp-HA and wild-type or mutant Egd2p-HA. TE and immunoprecipitates were analyzed with antibodies against HA or Rpl25p as indicated on the left. (E) Total protein extracts were prepared from _egd1_Δ cells expressing His6-ubiquitin and HA-tagged Egd1p, either wild type (Egd1WTp) or mutated in its ribosome-binding domain (Egd1RRK/AAAp). The ubiquitination assay was performed as in Figure 1A. (F) _egd1_Δ _egd2_Δ or _egd1_Δ _egd2_Δ _not4_Δ cells expressing HA-tagged Egd1RRK/AAAp were analyzed as in Figure 1B for the presence of the mutant Egd1p.
Figure 5.—
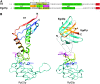
Comparison of the interaction of bacterial TF and yeast EGD with the ribosome. (A) Sequence alignments of D. radiodurans TF and S. cerevisiae Egd1p was derived from a multiple alignment that confirms the high conservation rate of several residues in this region over homologous sequences of TF and βNAC/Egd1p. The three parts of the ribosome-binding loop are indicated by numbers and orange, green, and purple, respectively. Identical, equivalent, and similar residues are shown with “|” (vertical line), “:” (colon), and “.” (period), respectively. Pale yellow indicates the conserved motifs known from the literature (K
ramer
et al. 2002; W
egrzyn
et al. 2006). The modeled regions are indicated in boldface type. Predicted α-helices and β-strands are in green and red, respectively. (B) TF is displayed colored from red to blue according to sequence position. The three parts of the ribosome-binding loop are indicated by numbers and orange, green and purple, respectively, and conserved residues interacting with D. radiodurans Rpl23p (G42, R44) or S. cerevisiae Rpl25p (G22, R24) are in green. Conserved P10 and E14 in Rpl23p and the corresponding P66 and E70 in Rpl25p are in blue. Egd1p and Egd2p are in orange and blue, respectively. K29 of Egd1p, the corresponding R50 of TF, and K30 of Egd1p, pointing to the surface of the ribosome, are in purple. From modeling of Egd1p and Egd2p, we can propose that EGD complex architecture is similar to the TF β-sheet structure although the exact position of α-helices (α1 and α2), indicated by dotted line in Egd1p, and the precise orientation of the Egd1p/Egd2p-interacting domains cannot be determined.
Figure 6.—
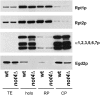
Egd2p interacts with the proteasome. Proteasome holoenzyme (holo), RP, and CP were purified from wild-type or _not4_Δ cell extracts by tandem affinity purification through the Rpn1p subunit. The presence of the indicated proteasome subunits was analyzed in TEs and in the different proteasome fractions by Western blot using antibodies against Rpt1p and Rpt2p or the α-subunits. α1, -2, -3, -5, -6, and 7 levels in TE were detectable only upon longer exposure times and were the same in wild type and _not4_Δ (data not shown). Copurification of Egd2p was analyzed by probing the blots with antibodies against Egd2p.
Similar articles
- The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex.
Panasenko O, Landrieux E, Feuermann M, Finka A, Paquet N, Collart MA. Panasenko O, et al. J Biol Chem. 2006 Oct 20;281(42):31389-98. doi: 10.1074/jbc.M604986200. Epub 2006 Aug 22. J Biol Chem. 2006. PMID: 16926149 - The yeast EGD2 gene encodes a homologue of the alpha NAC subunit of the human nascent-polypeptide-associated complex.
Shi X, Parthun MR, Jaehning JA. Shi X, et al. Gene. 1995 Nov 20;165(2):199-202. doi: 10.1016/0378-1119(95)00577-s. Gene. 1995. PMID: 8522175 - Functional Dissection of the Nascent Polypeptide-Associated Complex in Saccharomyces cerevisiae.
Ott AK, Locher L, Koch M, Deuerling E. Ott AK, et al. PLoS One. 2015 Nov 30;10(11):e0143457. doi: 10.1371/journal.pone.0143457. eCollection 2015. PLoS One. 2015. PMID: 26618777 Free PMC article. - The ribosome-bound quality control complex: from aberrant peptide clearance to proteostasis maintenance.
Defenouillère Q, Fromont-Racine M. Defenouillère Q, et al. Curr Genet. 2017 Dec;63(6):997-1005. doi: 10.1007/s00294-017-0708-5. Epub 2017 May 20. Curr Genet. 2017. PMID: 28528489 Review. - [Multifunctional protein complex NAC (nascent polypeptide associated complex].
Kogan GL, Gvozdev VA. Kogan GL, et al. Mol Biol (Mosk). 2014 Mar-Apr;48(2):223-31. Mol Biol (Mosk). 2014. PMID: 25850291 Review. Russian.
Cited by
- The ubiquitin-proteasome system of Saccharomyces cerevisiae.
Finley D, Ulrich HD, Sommer T, Kaiser P. Finley D, et al. Genetics. 2012 Oct;192(2):319-60. doi: 10.1534/genetics.112.140467. Genetics. 2012. PMID: 23028185 Free PMC article. Review. - Ubc4 and Not4 regulate steady-state levels of DNA polymerase-α to promote efficient and accurate DNA replication.
Haworth J, Alver RC, Anderson M, Bielinsky AK. Haworth J, et al. Mol Biol Cell. 2010 Sep 15;21(18):3205-19. doi: 10.1091/mbc.E09-06-0452. Epub 2010 Jul 21. Mol Biol Cell. 2010. PMID: 20660159 Free PMC article. - The essential Ubc4/Ubc5 function in yeast is HECT E3-dependent, and RING E3-dependent pathways require only monoubiquitin transfer by Ubc4.
Stoll KE, Brzovic PS, Davis TN, Klevit RE. Stoll KE, et al. J Biol Chem. 2011 Apr 29;286(17):15165-70. doi: 10.1074/jbc.M110.203968. Epub 2011 Feb 25. J Biol Chem. 2011. PMID: 21357418 Free PMC article. - Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its αNAC subunit.
Wang L, Zhang W, Wang L, Zhang XC, Li X, Rao Z. Wang L, et al. Protein Cell. 2010 Apr;1(4):406-416. doi: 10.1007/s13238-010-0049-3. Epub 2010 May 8. Protein Cell. 2010. PMID: 21203952 Free PMC article. - The Regulatory Properties of the Ccr4-Not Complex.
Chalabi Hagkarim N, Grand RJ. Chalabi Hagkarim N, et al. Cells. 2020 Oct 29;9(11):2379. doi: 10.3390/cells9112379. Cells. 2020. PMID: 33138308 Free PMC article. Review.
References
- Andersen, K. M., C. A. Semple and R. Hartmann-Petersen, 2007. Characterisation of the nascent polypeptide-associated complex in fission yeast. Mol. Biol. Rep. 34 275–281. - PubMed
- Beatrix, B., H. Sakai and M. Wiedmann, 2000. The alpha and beta subunit of the nascent polypeptide-associated complex have distinct functions. J. Biol. Chem. 275 37838–37845. - PubMed
- Bloss, T. A., E. S. Witze and J. H. Rothman, 2003. Suppression of CED-3-independent apoptosis by mitochondrial betaNAC in Caenorhabditis elegans. Nature 424 1066–1071. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases