HINT1 inhibits beta-catenin/TCF4, USF2 and NFkappaB activity in human hepatoma cells - PubMed (original) (raw)
HINT1 inhibits beta-catenin/TCF4, USF2 and NFkappaB activity in human hepatoma cells
Lin Wang et al. Int J Cancer. 2009.
Abstract
In this study we explored the relevance of Hint, a novel tumor suppressor gene, to human hepatoma. The human hepatoma cell lines Hep3B and HepG2 express very low levels of the HINT1 protein but the Huh7 cells express a relatively high level. In Hep3B and HepG2 cells, but not in Huh7 cells, the promoter region of Hint1 is partially methylated and treatment with 5-azadcdeoxycytidine increased expression of the HINT1 protein and Hint1 mRNA in Hep3B and HepG2 cells. Increased expression of HINT1 in HepG2 cells markedly inhibited their growth. It also inhibited the transcriptional activities of beta-catenin/TCF4, and USF2, and inhibited the expression of endogenous cyclin D1 and TGFbeta2. Furthermore, HINT1 co-immunoprecipitated with USF2 in extracts of Hep2 cells. HINT1 also inhibited NFkappaB transcription factor reporter activity and inhibited translocation of the endogenous p65 protein to the nucleus of HepG2 cells. Therefore, decreased expression of the Hint1 gene through epigenetic silencing may play a role in enhancing the growth of a subset of human hepatoma by increasing the expression of genes controlled by the transcription factors beta-catenin, USF2, and NFkappaB.
Figures
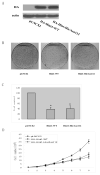
Figure 1. Levels of expression of the endogenous HINT1 protein and extent of methylation of the Hint1 promoter in human hepatoma cell lines
(A), Western blot analysis of HINT1 expression in extracts of the indicated cell lines. Actin was used as an internal loading control. HINT1 protein levels were quantified by densitometry and normalized to the corresponding level of actin. The expression ratios between the cell lines are presented in the lower panel. (B), Methylation Specific PCR (MSP). Total cellular DNA was extracted from the indicated cell lines, modified with bisulfite, and subjected to PCR with sets of specific PCR primers (U, unmethylated; M, methylated). PCR products were analyzed on 1% agarose gels and visualized under UV illumination. All assays ware repeated three times, and gave similar results. (C), Cells were treated with 5-Azadc (1μM) for 96 hours. Total RNA was isolated and semi-quantitative RT-PCR was done with specific Hint1 and GAPDH (internal loading control) primers. PCR products were analyzed on 1% agarose gels and visualized under UV illumination. (−) no treatment, (+) 5-Azadc. Expression ratios for Hint1 mRNA were calculated after normalization for GAPDH. All assays were repeated three times and gave similar results. (D), Cells were treated with 5-Azadc as described above, and HINT1 expression levels were detected by Western blot analysis. Actin was used as loading control. (−) no treatment, (+) 5-Azadc. Expression ratios for HINT1 protein were calculated after normalization for β-actin. Assays were done in duplicate and gave similar results.
Figure 2. Overexpression of HINT1 inhibits growth of HepG2 cells
The retrovirus shuttle plasmid pLNCX2 encoding HA-Hint1-WT, or HA-Hint1-His/Asn 112, or an empty control plasmid, was transfected into the PT67 packaging cell line. After incubation for 48h the retrovirus containing medium was harvested. HepG2 cells were then infected with the indicated retroviruses for 24 h at a multiplicity of infection of 800 and then incubated in fresh medium for 72 h. (A), Total cell lysates were prepared and Western blots performed with an Anti-HA antibody. (B) and (C), Colony formation assay. The infected cells were grown in the presence of the selection agent G418 for 3 weeks. The colonies were then fixed, stained with Giemsa solution, and counted. The photographs (B) display representative plates and the bar graph (C) displays the percent inhibition of colony formation. The results are the means and standard deviations (S.D.) of triplicate assays. The asterisk * indicates significant inhibition (p<0.01). (D) Growth curves. Cells were cultured in 24 well plates, infected with the above described retrovirus-containing medium and the number of cells in replicate wells was counted every day for the subsequent 8 days. The data indicate the mean values and S.D.s of triplicate assays. Similar results were obtained in a repeat assay.
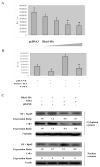
Figure 3. HINT1 inhibits β-catenin/TCF4 transcription factor activity and cyclin D1 expression in HepG2 cells
(A) The Topflash luciferase reporter and CMV-β-gal reporter plasmid DNAs were transfected into the HepG2 cells. As indicated, increasing amounts of pHA-Hint1 plasmid DNA (0, 0.5, 1.0, 1.5, 2.0/well) were also co-transfected, and pcDNA was added, as needed, to achieve the same total amount of plasmid DNA in each assay. Luciferase activity was assayed at 36 h post-transfection and normalized for β-gal activity. All assays were done in triplicate and the S.D.s are shown. Similar results were obtained in a repeat study. No significant luciferase activity was obtained with the negative control Fopflash luciferase reporter. (B) The cyclin D1 reporter CD1 (-1745)-LUC and CMV-β-gal reporter plasmid DNAs were co-transfected into HepG2 cells, with our without increasing amounts of Hint1 plasmid DNA and luciferase activity assayed 36 hours post transfection as described in Fig. 3A. (C) the cyclin D1 reporter assays were done as in (B), with co-transfected Hint1-HA or wild type β-catenin plasmid DNAs, as indicated. (D) HepG cells were infected with the indicated retrovirus containing medium as described in Fig. 2A. At 72 hours post infection, whole cell protein extracts were prepared and the levels of cyclin D1 protein expression were determined by Western blot analysis. Actin was used as loading control. Expression ratios for cyclin D1 protein were calculated after normalization for β-actin. (E) HepG2 cells were treated as in (D) and at 36 hours cellular RNA was extracted and semi-quantitative RT-PCR was performed using specific primer sets for cyclin D1 and GAPDH. RT-PCR products were analyzed on 1% agarose gels and visualized under UV illumination. Expression ratios for cyclin D1 mRNA were calculated after normalization for GAPDH. All assays were repeated twice and gave similar results.
Figure 4. HINT1 inhibits USF2/E-box related transcription factor activity and TGFβ2 expression in HepG2 cells
(A), The p-cathepsin K-LUC or pE-box-LUC, and CMV-β-gal reporter plasmid DNAs, were co-transfected into HepG2 cells. pHANE, pHA-Hint1 and pUSF2 plasmid DNAs were also co-transfected, as indicated. Luciferase activity was assayed 36 hours later and corrected for β-gal activity, as described in Fig. 3A. All assays were done in triplicate and a repeat study gave similar results. (B), HepG2 cells were transfected with pCMV-USF2 or pHA-Hint1 plasmid DNAs as indicated. After 48 hours, whole cell protein extracts were prepared and were immunoprecipitated with an anti-HA monoclonal affinity gel (left panel) or an anti-USF2 polyclonal antibody together with protein G plus-agarose (right panel), at 4°C for 18 hours. The immunoprecipitates were washed with cold lysis buffer five times, and then analyzed on Western blots with an anti-USF2 polyclonal antibody or an anti-HA monoclonal antibody to detect the USF2 or the HA tagged HINT1 proteins in the immunocomplexes and in the whole cell lysate (bottom panel), as indicated. (C), HepG2 cells were transfected with wild type (p77-CAT) or mutant (p40-CAT, p77E-CAT) TGF-β2 promoter/CAT reporter plasmid DNAs together with the CMV-β-gal reporter plasmids DNA. At 36 hour post transfection, whole cell extracts were prepared, and CAT activity was assayed and normalized for β-gal activity. The results are expressed as the CAT activities of the pβ2-77 or pβ2-77E relative to the activity of the pβ2-40 construct. All assays were done in triplicate and a repeat study gave similar results. (D), pHA-Hint1 or the pCMV-USF2 plasmid DNAs were transfected into HepG2 cells, as indicated. At 48 hours post transfection, whole cell RNA was extracted, and semi-quantitative RT-PCR was performed using specific TGF β2 and GAPDH primer sets, as described in Fig. 4D. Assays were repeated twice and gave similar results.
Figure 5. HINT1 inhibits NF-κB transcription factor activity in HepG2 cells
(A) and (B), pNF-κB-Luc (400 ng/well) and the CMV-β-gal reporter control plasmid DNAs, together with pHA-Hint1 (0, 0.5, 1.0, 1.5, 2.0 μg) in (B) or pHA-Hint1 (1.0 μg) with our without CMV-USF2 (1.0 μg) plasmid DNAs, as indicated, were transfected into the HepG2 cells. Thirty-six hours later luciferase activity was assayed as described in Fig. 3A. (C), pHA-Hint1 or the CMV-USF2 plasmid DNAs were transfected into HepG2 cells, as indicated. At 48 hours post transfection, cytoplasmic and nuclear proteins were extracted, and each fraction was analyzed by Western blots to detect the expression levels of p65, IκBα, β-tubulin and β-actin. The latter two proteins were used as loading controls for the cytoplasmic and nuclear extracts, respectively. Expression ratios were calculated after normalization for β-tubulin and β-actin. Assays were repeated three times and gave similar results.
Similar articles
- Hint1 Up-Regulates IκBα by Targeting the β-TrCP Subunit of SCF E3 Ligase in Human Hepatocellular Carcinoma Cells.
Shi Z, Wu X, Ke Y, Wang L. Shi Z, et al. Dig Dis Sci. 2016 Mar;61(3):785-94. doi: 10.1007/s10620-015-3927-y. Epub 2015 Oct 31. Dig Dis Sci. 2016. PMID: 26520111 - SOX10 is a novel oncogene in hepatocellular carcinoma through Wnt/β-catenin/TCF4 cascade.
Zhou D, Bai F, Zhang X, Hu M, Zhao G, Zhao Z, Liu R. Zhou D, et al. Tumour Biol. 2014 Oct;35(10):9935-40. doi: 10.1007/s13277-014-1893-1. Epub 2014 Jul 8. Tumour Biol. 2014. PMID: 25001176 - Beta-catenin regulates vitamin C biosynthesis and cell survival in murine liver.
Nejak-Bowen KN, Zeng G, Tan X, Cieply B, Monga SP. Nejak-Bowen KN, et al. J Biol Chem. 2009 Oct 9;284(41):28115-28127. doi: 10.1074/jbc.M109.047258. Epub 2009 Aug 18. J Biol Chem. 2009. PMID: 19690176 Free PMC article. - [HINT1--a novel tumor suppressor protein of the HIT superfamily].
Ozga M. Ozga M. Postepy Biochem. 2010;56(1):55-60. Postepy Biochem. 2010. PMID: 20499681 Review. Polish. - Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma.
Ding SL, Yang ZW, Wang J, Zhang XL, Chen XM, Lu FM. Ding SL, et al. World J Gastroenterol. 2015 May 28;21(20):6317-28. doi: 10.3748/wjg.v21.i20.6317. World J Gastroenterol. 2015. PMID: 26034368 Free PMC article. Review.
Cited by
- NUDT2 Disruption Elevates Diadenosine Tetraphosphate (Ap4A) and Down-Regulates Immune Response and Cancer Promotion Genes.
Marriott AS, Vasieva O, Fang Y, Copeland NA, McLennan AG, Jones NJ. Marriott AS, et al. PLoS One. 2016 May 4;11(5):e0154674. doi: 10.1371/journal.pone.0154674. eCollection 2016. PLoS One. 2016. PMID: 27144453 Free PMC article. - The Axonal Motor Neuropathy-Related HINT1 Protein Is a Zinc- and Calmodulin-Regulated Cysteine SUMO Protease.
Cortés-Montero E, Rodríguez-Muñoz M, Sánchez-Blázquez P, Garzón J. Cortés-Montero E, et al. Antioxid Redox Signal. 2019 Sep 1;31(7):503-520. doi: 10.1089/ars.2019.7724. Epub 2019 Jun 24. Antioxid Redox Signal. 2019. PMID: 31088288 Free PMC article. - Expression, purification, crystallization and preliminary X-ray crystallographic analysis of human histidine triad nucleotide-binding protein 2 (hHINT2).
Dolot R, Włodarczyk A, Bujacz GD, Nawrot B. Dolot R, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Jul;69(Pt 7):783-7. doi: 10.1107/S1744309113015200. Epub 2013 Jun 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23832208 Free PMC article. - Metabolic Reprogramming and the Recovery of Physiological Functionality in 3D Cultures in Micro-Bioreactors.
Wrzesinski K, Fey SJ. Wrzesinski K, et al. Bioengineering (Basel). 2018 Mar 7;5(1):22. doi: 10.3390/bioengineering5010022. Bioengineering (Basel). 2018. PMID: 29518979 Free PMC article. - Taraxasterol suppresses the growth of human liver cancer by upregulating Hint1 expression.
Bao T, Ke Y, Wang Y, Wang W, Li Y, Wang Y, Kui X, Zhou Q, Zhou H, Zhang C, Zhou D, Wang L, Xiao C. Bao T, et al. J Mol Med (Berl). 2018 Jul;96(7):661-672. doi: 10.1007/s00109-018-1652-7. Epub 2018 May 27. J Mol Med (Berl). 2018. PMID: 29806073
References
- Li H, Zhang Y, Su T, Santella RM, Weinstein IB. Hint1 is a haplo-insufficient tumor suppressor in mice. Oncogene. 2006;25:713–21. - PubMed
- Wang L, Zhang Y, Li H, Xu Z, Santella RM, Weinstein IB. Hint1 inhibits growth and activator protein-1 activity in human colon cancer cells. Cancer Res. 2007;67:4700–8. - PubMed
- Korsisaari N, Makela TP. Interactions of Cdk7 and Kin28 with Hint/PKCI-1 and Hnt1 histidine triad proteins. J Biol Chem. 2000;275:34837–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 ES009089/ES/NIEHS NIH HHS/United States
- P30 ES009089-10/ES/NIEHS NIH HHS/United States
- R01 ES005116/ES/NIEHS NIH HHS/United States
- ES09089/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials