Biochemical analyses of human IST1 and its function in cytokinesis - PubMed (original) (raw)
Biochemical analyses of human IST1 and its function in cytokinesis
Monika Bajorek et al. Mol Biol Cell. 2009 Mar.
Abstract
The newly described yeast endosomal sorting complexes required for transport (ESCRT) protein increased sodium tolerance-1 (Ist1p) binds the late-acting ESCRT proteins Did2p/charged MVB protein (CHMP) 1 and Vps4p and exhibits synthetic vacuolar protein sorting defects when combined with mutations in the Vta1p/LIP5-Vps60p/CHMP5 complex. Here, we report that human IST1 also functions in the ESCRT pathway and is required for efficient abscission during HeLa cell cytokinesis. IST1 binding interactions with VPS4, CHMP1, LIP5, and ESCRT-I were characterized, and the IST1-VPS4 interaction was investigated in detail. Mutational and NMR spectroscopic studies revealed that the IST1 terminus contains two distinct MIT interacting motifs (MIM1 and MIM2) that wrap around and bind in different groves of the MIT helical bundle. IST1, CHMP1, and VPS4 were recruited to the midbodies of dividing cells, and depleting either IST1 or CHMP1 proteins blocked VPS4 recruitment and abscission. In contrast, IST1 depletion did not inhibit human immunodeficiency virus-1 budding. Thus, IST1 and CHMP1 act together to recruit and modulate specific VPS4 activities required during the final stages of cell division.
Figures
Figure 1.
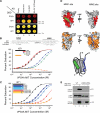
IST1 binding interactions. (A) Yeast two-hybrid interactions of IST1 with other human ESCRT pathway proteins. IST1-AD fusions (columns 2, IST1) or control AD constructs (columns 1, Empty) were coexpressed with control DBD constructs (row 1, Empty) or ESCRT protein–DBD fusions (rows 2–26) and tested for positive yeast two-hybrid interactions (left) or cotransformation (control, right). IST1 showed positive interactions with CHMP1A, CHMP1B, VPS4A, VPS4B, and LIP5 in this assay. (B) IST1 binds ESCRT-I. Left, ESCRT-I coprecipitations with an empty vector control (lane 1) or One-STrEP-FLAG(OSF)-IST1 (lane 2). All four ESCRT-I subunits were coexpressed with N-terminal Myc epitope tags in these experiments. Right, binding of pure recombinant ESCRT-I complexes to GST-IST1 in E. coli extracts. Top right, Coomassie-stained SDS-PAGE gel showing pure recombinant ESCRT-I (lane 1), GST or GST-IST1 control extracts (lanes 2 and 3), and ESCRT-I pull-down reactions (lanes 4 and 5). Bottom right, Western blot showing matrix binding levels of the FLAG-TSG101/ESCRT-I subunit in the reactions corresponding to lanes 2–5 of the top panel. (C) IST1 binds CHMP1A. Left, Myc-CHMP1A coprecipitations with empty vector controls (lanes 1, 3, and 5), OSF-IST1 (lane 2), OSF-IST11-189 (lane 4), or OSF-IST1190-366 (lane 6). Right, Myc-CHMP1A (lanes 1 and 2) and Myc-CHMP1AL191A/L184A (lanes 3 and 4) coprecipitations with empty vector controls (lanes 1 and 3) and OSF-IST1 (lanes 2 and 4). (D) IST1 binds LIP5. Left, Myc-LIP5 coprecipitations with empty vector controls (lanes 1, 3, and 5), or OSF-IST1 (lane 2), OSF-IST11-189 (lane 4) or OSF-IST190-366 (lane 6). Right, endogenous IST1 coprecipitations with empty vector controls (lanes 1, 3, and 5), OSF-LIP5 (lane 2), OSF-LIP51-186 (lane 4), or OSF-LIP5197-307 (lane 6). (E) Summary of IST1 protein interactions. Domain abbreviations: VSL, _V_ta1/_S_BP1/_L_IP5 domain; UEV, _U_biquitin _E_2 _V_ariant domain; PRD, proline-_r_ich _d_omain; NTD/CTD, N- and C-terminal domains; β, β-domain; AAA ATPase, _A_TPases _a_ssociated with a variety of cellular _a_ctivities.
Figure 2.
IST1 Interacts with VPS4A MIT. (A) Yeast two-hybrid interactions between IST1 and designated VPS4A constructs. Two-hybrid interactions (top) or cotransformation controls (bottom) are shown. Note that IST1 showed a positive two-hybrid interaction with the VPS4A MIT domain but not the VPS4A ATPase cassette (compare lanes 2 and 3 and 4). (B) IST1 has two distinct MIM elements. Top, alignment of IST1321-366 (top) with CHMP6 MIM2 and CHMP1A MIM1 (middle), and with MIM consensus sequences (bottom). Bottom, biosensor binding isotherms showing pure VPS4A MIT binding to GST-IST1 proteins captured from E. coli extracts. Estimated dissociation constants (micromolar) for VPS4A MIT binding were as follows: IST1, 1.1 (black); IST1303-366, 0.8 (red); IST1323-339, 12.4 (blue); IST1340-366, 26 (green); and IST11-189, >1500 (orange). Background binding of VPS4A MIT to control GST surfaces was negligible (data not shown). (C) Mutational analyses of IST1 MIM1 and MIM2. Biosensor binding isotherms showing purified VPS4A MIT binding to GST-IST1 proteins captured from E. coli extracts. Estimated dissociation constants (micromolar) for VPS4A MIT binding were as follows: IST1, 1.1 (black); IST1L325D, 8; IST1L328D, 16; IST1L355A, 17; IST1L362A, 9 (single mutants are in blue and purple shades); IST1L325D/L355A, 88; IST1L328D/L355A, 502; and IST1L328/L362, 432 (double mutants are in red and orange shades). (D) NMR chemical shift mapping of the IST1 MIM1 and MIM2 binding sites on VPS4A MIT. Figures show residues shifted by binding of IST1 MIM1 mapped onto space filling models of the VPS4A MIT domain. Binding modes of the structurally characterized CHMP1A MIM1 (top; green) and CHMP6 (middle; blue) elements are shown for reference. The bottom panel shows a structure-based cartoon model of the VPS4A MIT-IST1321-366 complex, with the three VPS4A MIT helices depicted in gray, and the IST1 MIM1 and MIM2 elements depicted in green and blue, respectively. (E) LIP5 binds the MIM1/MIM2 region of IST1. Wild-type (WT) Myc-IST1 (lanes 1 and 2) or Myc-IST1L328D/L355A (lanes 3 and 4) coprecipitations with empty vector controls (lanes 1 and 3) or with OSF-LIP5 (lanes 2 and 4).
Figure 3.
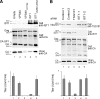
IST1 overexpression inhibits HIV budding but IST1 depletion does not. (A) IST1 overexpression reduces HIV-1 release and titer. Protein levels (Western blots) and vector titers (infectious units per milliliter; bottom) upon coexpression of an HIV-1 vector with an empty vector (lane 1, negative control) or with vectors expressing GFP-VPS4A (lane 2, negative control), GFP-Vps4AK173Q (lane 3, positive control), IST1 (lane 4), or IST1L328D/L355A (lane 5). Western blots show intracellular IST1 (top, anti-IST1), intracellular HIV Gag proteins (middle, anti-CA), and virion-associated CA proteins (bottom, anti-CA). (B) IST1 depletion does not reduce HIV-1 vector release or titer. Protein levels (Western blots) and vector titers (bottom) upon coexpression of an HIV-1 vector with two irrelevant siRNAs (lanes 1 and 2, negative controls), an siRNA against TSG101 (positive control, lane 3), or two different siRNAs against IST1 (lanes 4 and 5). Western blots show intracellular TSG101 (panel 1, anti-TSG101), intracellular IST1 (panel 2, anti-IST1), intracellular HIV Gag proteins (panel 3, anti-CA), and virion-associated CA proteins (panel 4, anti-CA).
Figure 4.
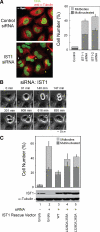
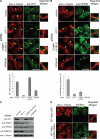
IST1 is required for efficient cytokinesis. (A) IST1 depletion causes cells to arrest during cytokinesis and to accumulate multiple nuclei. Left, confocal immunofluorescence images of HeLa cells treated with a control siRNA (TOP) or an siRNA against IST1 (BOTTOM). Microtubules (anti-α-Tubulin, red) and nuclei (SYTOX-green) were stained for reference; the white arrowhead highlights a multinuclear cell, and yellow arrowheads highlight cells arrested in cytokinesis. Right, Quantification of cells with multiple nuclei (dark bars) or with midbodies (light bars) after treatment with the irrelevant control siRNA or with either of the two different siRNAs against IST1. (B) IST1 depletion blocks the abscission stage of cytokinesis. Time-lapse, phase contrast images of a dividing cell depleted of IST1. The figure shows a cell depleted of IST1 that arrests late in cytokinesis (t = 147 min; elapsed times are provided above the panels), remains tethered via a midbody for an extended period (t = 147–609 min), and then eventually recoalesces to form a multinucleated cell (t = 616 min). A full movie is provided in Supplemental Figure S3. (C) Rescue of cytokinesis defects with siRNA-resistant IST1 constructs. Percentages of cells with multiple nuclei (dark bars) or with midbodies (light bars) after depletion of endogenous IST1 (lane 2) or after depletion of IST1 and re-expression of exogenous wild-type IST1 (lane 3) or IST1 constructs with two different sets of MIM1/MIM2 mutations (lanes 4 and 5). Lane 1 shows cells that were treated with a control siRNA and cotransfected with an empty expression vector. Western blots (below) show cellular IST1 levels and anti-α-tubulin loading controls.
Figure 5.
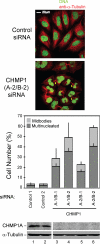
CHMP1 proteins are required for efficient cytokinesis. Simultaneous depletion of CHMP1A and CHMP1B causes cells to arrest during cytokinesis and to accumulate multiple nuclei. Top, confocal immunofluorescence images are analogous to those shown in Figure 4A, except that cells in the bottom panel were treated with siRNAs targeting both CHMP1A and CHMP1B. Bottom, quantification of cells with multiple nuclei (dark bars) or with midbodies (light bars). These experiments are analogous to those shown in Figure 4C, except that 1) cells were treated with different pairs of siRNAs that targeted both CHMP1A and CHMP1B, and 2) only CHMP1A protein levels were examined because CHMP1B antibodies are not yet available.
Figure 6.
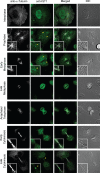
IST1 localization changes through the cell cycle. Representative images showing localization of endogenous HeLa IST1 (anti-IST1, green, column 2) at different stages of the cell cycle. Magnified insets show IST1 concentrated at the microtubule-organizing center/centrosome during prophase and early metaphase (rows 2 and 3) and at the midbody late in cytokinesis (row 7). Corresponding images show the localization of microtubules (anti-α-Tubulin, white, column 1), merged images (column 3), and differential interference contrast (DIC) images (column 4).
Figure 7.
Requirements for the midbody localization of IST1 and VPS4. (A) Requirements for IST1 localization to the Flemming bodies of dividing cells. Doubled-labeled confocal immunofluorescence images showing the localization of endogenous IST1 in dividing HeLa cells (anti-IST1, green, column 2); microtubules (anti-α-tubulin, red, column 1); and merged, magnified views (column 3). IST1 localization patterns are shown in the presence of a control siRNA (row 1) and in cells depleted of IST1 (row 2), CHMP1A and CHMP1B (row 3), or VPS4A and VPS4B (row 4). Percentages of midbodies with observable IST1 under the different conditions are quantified below. Note that IST1 concentrated in double ring structures within midbodies (arrowheads) except when IST1 itself was depleted. (B) Requirements for VPS4B localization to the Flemming bodies of dividing cells. Doubled-labeled confocal immunofluorescence images showing localization of endogenous VPS4B in dividing HeLa cells (anti-VPS4B, green, column 2), microtubules (anti-α-tubulin, red, column 1), and merged, magnified views (column 3). Percentages of midbodies with observable VPS4B under the different conditions are quantified below. Note that VPS4B was not efficiently recruited to midbodies (arrowheads) when either IST1, CHMP1, or VPS4 proteins were depleted. (C) Western blots showing the relative levels of IST1, VPS4, CHMP1, and α-Tubulin proteins under the different experimental conditions shown in A and B. Note that CHMP1A/B depletion reduced VPS4A levels and that depletion of VPS4A/B reduced CHMP1A to undetectable levels. (D) IST1 mutations that inhibit VPS4 MIT binding do not inhibit midbody localization. Both wild-type IST1-Myc protein (row 1, column 2) and an IST1 mutant that lacks the ability to bind VPS4 MIT domains (IST1L328D,L355A-Myc, row 2, column 2) localized to the midbodies of dividing cells. Microtubule localization (anti-α-tubulin, red, column 1) and merged, magnified views (column 3) are also shown for reference.
Similar articles
- MITD1 is recruited to midbodies by ESCRT-III and participates in cytokinesis.
Lee S, Chang J, Renvoisé B, Tipirneni A, Yang S, Blackstone C. Lee S, et al. Mol Biol Cell. 2012 Nov;23(22):4347-61. doi: 10.1091/mbc.E12-04-0292. Epub 2012 Sep 26. Mol Biol Cell. 2012. PMID: 23015756 Free PMC article. - Interactions of the human LIP5 regulatory protein with endosomal sorting complexes required for transport.
Skalicky JJ, Arii J, Wenzel DM, Stubblefield WM, Katsuyama A, Uter NT, Bajorek M, Myszka DG, Sundquist WI. Skalicky JJ, et al. J Biol Chem. 2012 Dec 21;287(52):43910-26. doi: 10.1074/jbc.M112.417899. Epub 2012 Oct 26. J Biol Chem. 2012. PMID: 23105106 Free PMC article. - The regulation of Endosomal Sorting Complex Required for Transport and accessory proteins in multivesicular body sorting and enveloped viral budding - An overview.
Ahmed I, Akram Z, Iqbal HMN, Munn AL. Ahmed I, et al. Int J Biol Macromol. 2019 Apr 15;127:1-11. doi: 10.1016/j.ijbiomac.2019.01.015. Epub 2019 Jan 4. Int J Biol Macromol. 2019. PMID: 30615963 Review. - Regulation of Vps4 during MVB sorting and cytokinesis.
Babst M, Davies BA, Katzmann DJ. Babst M, et al. Traffic. 2011 Oct;12(10):1298-305. doi: 10.1111/j.1600-0854.2011.01230.x. Epub 2011 Jul 7. Traffic. 2011. PMID: 21658171 Free PMC article. Review.
Cited by
- Overexpression of OLC1 promotes tumorigenesis of human esophageal squamous cell carcinoma.
Li X, Suo J, Shao S, Xue L, Chen W, Dong L, Shi J, Fu M, Lu N, Zhan Q, Tong T. Li X, et al. PLoS One. 2014 Mar 7;9(3):e90958. doi: 10.1371/journal.pone.0090958. eCollection 2014. PLoS One. 2014. PMID: 24608342 Free PMC article. - Spastin MIT Domain Disease-Associated Mutations Disrupt Lysosomal Function.
Allison R, Edgar JR, Reid E. Allison R, et al. Front Neurosci. 2019 Nov 8;13:1179. doi: 10.3389/fnins.2019.01179. eCollection 2019. Front Neurosci. 2019. PMID: 31787869 Free PMC article. - ESCRT requirements for EIAV budding.
Sandrin V, Sundquist WI. Sandrin V, et al. Retrovirology. 2013 Oct 9;10:104. doi: 10.1186/1742-4690-10-104. Retrovirology. 2013. PMID: 24107264 Free PMC article. - Identification of abscission checkpoint bodies as structures that regulate ESCRT factors to control abscission timing.
Strohacker LK, Mackay DR, Whitney MA, Couldwell GC, Sundquist WI, Ullman KS. Strohacker LK, et al. Elife. 2021 Aug 4;10:e63743. doi: 10.7554/eLife.63743. Elife. 2021. PMID: 34346309 Free PMC article. - Meiotic Clade AAA ATPases: Protein Polymer Disassembly Machines.
Monroe N, Hill CP. Monroe N, et al. J Mol Biol. 2016 May 8;428(9 Pt B):1897-911. doi: 10.1016/j.jmb.2015.11.004. Epub 2015 Nov 10. J Mol Biol. 2016. PMID: 26555750 Free PMC article. Review.
References
- Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., Katzmann D. J. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell. 2008;14:50–61. - PubMed
- Babst M., Katzmann D., Estepa-Sabal E., Meerloo T., Emr S. Escrt-III. An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 2002;3:271–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous