Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission - PubMed (original) (raw)
Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission
Ruben K Dagda et al. J Biol Chem. 2009.
Abstract
Mitochondrial dysregulation is strongly implicated in Parkinson disease. Mutations in PTEN-induced kinase 1 (PINK1) are associated with familial parkinsonism and neuropsychiatric disorders. Although overexpressed PINK1 is neuroprotective, less is known about neuronal responses to loss of PINK1 function. We found that stable knockdown of PINK1 induced mitochondrial fragmentation and autophagy in SH-SY5Y cells, which was reversed by the reintroduction of an RNA interference (RNAi)-resistant plasmid for PINK1. Moreover, stable or transient overexpression of wild-type PINK1 increased mitochondrial interconnectivity and suppressed toxin-induced autophagy/mitophagy. Mitochondrial oxidant production played an essential role in triggering mitochondrial fragmentation and autophagy in PINK1 shRNA lines. Autophagy/mitophagy served a protective role in limiting cell death, and overexpressing Parkin further enhanced this protective mitophagic response. The dominant negative Drp1 mutant inhibited both fission and mitophagy in PINK1-deficient cells. Interestingly, RNAi knockdown of autophagy proteins Atg7 and LC3/Atg8 also decreased mitochondrial fragmentation without affecting oxidative stress, suggesting active involvement of autophagy in morphologic remodeling of mitochondria for clearance. To summarize, loss of PINK1 function elicits oxidative stress and mitochondrial turnover coordinated by the autophagic and fission/fusion machineries. Furthermore, PINK1 and Parkin may cooperate through different mechanisms to maintain mitochondrial homeostasis.
Figures
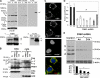
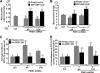
FIGURE 1.
Characterization of PINK1-overexpressing and knockdown cell lines. A, cell lysates from stable pRec vector control SH-SY5Y cell line (C), a PINK1 knockdown stable cell line (A14), and a PINK1-overexpressing clonal cell line (#24) were resolved on a 5-15% ammediol-buffered gel and immunoblotted for PINK1 with C8830 antisera in the presence (middle panel) or absence (left panel) of blocking PINK1 peptide (100 μg/ml for 1 h). The blot was reprobed for FLAG tag (right panel) and for β-actin as loading control. Inset 1 shows a longer exposure of the film to better visualize full-length endogenous PINK1 relative to the overexpressed PINK1-3xFlag. Immunoreactive bands include full-length recombinant PINK1-3xFlag (band _a_′, ∼69 kDa), endogenous full-length (band a, ∼66 kDa) and a major processed band (band b, ∼50 kDa). One or two lower molecular mass processed forms (<39 kDa) were also observed. A processed form of PINK1-3xFlag was sometimes observed (∼60 kDa, supplemental Fig. S8); Inset 2 shows the accentuation of this processed band in cells treated with the proteasome inhibitor clasto-lactacystin β-lactone (band_a_″). Smaller processed PINK1 bands likely reflect further processing, potentially at the C-terminal end, as no smaller FLAG bands are observed. B, Western blots of mitochondrial (mito) and cytoplasmic (cyto) fractions derived from parental SH-SY5Y cells (W), a stable vector line (Ctrl), and PINK1-3xFlag clone #24 resolved on a 10% Tris-glycine gel and immunoblotted for FLAG, endogenous PINK1, mitochondrial P60, and lactate dehydrogenase (LDH). Similar results were obtained for clone 9 (data not shown). Asterisk, indicates a nonspecific band. Full-length recombinant PINK1-3xFlag (band_a_′), endogenous full-length (band a), and processed (band b) PINK1 are observed. C, epifluorescence image of SH-SY5Y cells double labeled for endogenous PINK1 (green) and 60-kDa human mitochondrial antigen (red) and counterstained for nuclei with DAPI (blue). D, PINK1 mRNA levels in clonal SH-SY5Y lines that express shRNA directed against the N-terminal (A series clones) or middle (D series clones) regions of PINK1. Data are normalized to β-actin mRNA as relative quantification values ± max and min. A control clone generated using pRS control vector (V series clones) served as the reference value. *, p < 0.001 versus V17. E, PINK1 immunoblot showing reduction of endogenous PINK1 in stable PINK1shRNA cell lines compared with a vector control cell lines, with β-actin serving as the loading control. The 50-kDa processed band (b) and two smaller processed bands (c and d) are observed in this blot (see supplemental Fig. S7 for uncropped version of the ECL film). Densitometry data (compiled from three independent experiments for each line) is shown below (*, p < 0.01 versus control cell line).
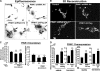
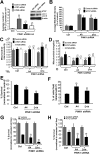
FIGURE 2.
Ultrastructural analysis of PINK1 knockdown and overexpressing cell lines. A-D, electron micrographs of stable cell lines that express empty shRNA vector control (A), A and D series PINK1 shRNAs (B and C), or PINK1-3xFlag (D). m, mitochondria; av, autophagic vacuoles; ly, lysosomes;n, nuclei. Inset, enlarged mitochondria showing dense cristae (scale bars:2 μm). E, the average number of cristae folds/μm of mitochondria ( , p < 0.001_versus_ vector control; mean ± S.E., n = 80-150 mitochondria/condition). F, average number of AVs (autophagosomes and autolysosomes) quantified per cell. (
, p < 0.001_versus_ vector control; mean ± S.E., n = 80-150 mitochondria/condition). F, average number of AVs (autophagosomes and autolysosomes) quantified per cell. ( , p < 0.05_versus_ vector control; mean ± S.E., n = 30 cells/condition).
, p < 0.05_versus_ vector control; mean ± S.E., n = 30 cells/condition).
FIGURE 3.
Knockdown or overexpression of PINK1 alters mitochondrial morphology. A, epifluorescence images of mitochondria labeled by transient transfection of mito-GFP in vector control, stable PINK1 shRNA lines, and a stable PINK1-overexpressing clone. Scale bar:20 μm.B, three-dimensional reconstruction of Z-stacks of mitochondria labeled with mito-GFP in vector control, PINK1 shRNA clones, and a PINK1-overexpressing clone. C, quantitative image analysis of mitochondrial interconnectivity and mitochondrial elongation applied to empty vector control and PINK1shRNA clonal cells (representative of three experiments;  , p < 0.02 versus control (Ctrl); mean ± S.E.,n = 25-35 cells). D, quantification of mitochondrial interconnectivity and mitochondrial elongation in a PINK1-3xFlag-overexpressing cell line and in SH-SY5Y cells transiently transfected with PINK1-WT-V5 (
, p < 0.02 versus control (Ctrl); mean ± S.E.,n = 25-35 cells). D, quantification of mitochondrial interconnectivity and mitochondrial elongation in a PINK1-3xFlag-overexpressing cell line and in SH-SY5Y cells transiently transfected with PINK1-WT-V5 ( , p < 0.05_versus_ stable controls or empty vector; mean ± S.E.,n = 25-35 cells/condition, three independent experiments).
, p < 0.05_versus_ stable controls or empty vector; mean ± S.E.,n = 25-35 cells/condition, three independent experiments).
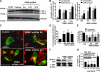
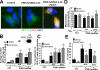
FIGURE 4.
Knockdown of PINK1 increases macroautophagy and mitophagy. A, LC3 Western blot demonstrating increase in LC3-II to β-actin ratios indicative of elevated autophagy in PINK1shRNA clones compared with a vector control clone or parental SH-SY5Y cells. B, the average number of GFP-LC3 puncta quantified per cell in a vector control line and two PINK1 shRNA clonal cell lines transiently expressing GFP-LC3 (marker of early AVs (left graph)) or RFP-LC3 (marker of later AVs (right graph)) in the presence (black bars) or absence (gray bars) of bafilomycin to inhibit lysosomal degradation ( , p < 0.01_versus_ control cell line (Ctrl); n = 20-40 cells quantified/condition;
, p < 0.01_versus_ control cell line (Ctrl); n = 20-40 cells quantified/condition;  ,p < 0.01 versus PINK1shRNA line in the absence of bafilomycin; n = 30-40 cells analyzed/condition). C, confocal microscopy of MitoTracker Red-labeled mitochondria in control and three PINK1shRNA lines transiently expressing GFP-LC3 (green).Scale bar:20 μm. D, quantitative analysis of GFP-LC3/MitoTracker colocalization as index of mitophagy in vector control and three PINK1 shRNA clonal lines. Bar graph shows means ± S.E. from 45-90 cells/condition compiled from four experiments (
,p < 0.01 versus PINK1shRNA line in the absence of bafilomycin; n = 30-40 cells analyzed/condition). C, confocal microscopy of MitoTracker Red-labeled mitochondria in control and three PINK1shRNA lines transiently expressing GFP-LC3 (green).Scale bar:20 μm. D, quantitative analysis of GFP-LC3/MitoTracker colocalization as index of mitophagy in vector control and three PINK1 shRNA clonal lines. Bar graph shows means ± S.E. from 45-90 cells/condition compiled from four experiments ( , p < 0.001_versus_ control cell line; n = 25-30 cells/condition).E, computer-aided quantification of the percent of cellular area occupied by mitochondria in control and PINK1 shRNA cells in the presence or absence of bafilomycin A. Cells were transiently transfected with mitochondrially targeted GFP and untargeted RFP to label the cellular perimeters. (
, p < 0.001_versus_ control cell line; n = 25-30 cells/condition).E, computer-aided quantification of the percent of cellular area occupied by mitochondria in control and PINK1 shRNA cells in the presence or absence of bafilomycin A. Cells were transiently transfected with mitochondrially targeted GFP and untargeted RFP to label the cellular perimeters. ( , p < 0.05 versus control cell line;
, p < 0.05 versus control cell line; , p < 0.05_versus_ untreated PINK1shRNA cell lines; n = 25-35 cells analyzed/condition) F, protein levels of mitochondrial pyruvate dehydrogenase (PDH), reprobed for total ERK1/2 as a loading control.G, computer-aided quantification of the integrated intensity of immunoreactive bands specific for human mitochondrial antigen of 60 kDa (mito-P60) relative to total ERK levels in a control cell line, three PINK1shRNA cell lines, and a PINK1-3x-Flag-overexpressing line. The bar graph shows means ± S.E. compiled from 3-4 Western blot experiments for each cell line (
, p < 0.05_versus_ untreated PINK1shRNA cell lines; n = 25-35 cells analyzed/condition) F, protein levels of mitochondrial pyruvate dehydrogenase (PDH), reprobed for total ERK1/2 as a loading control.G, computer-aided quantification of the integrated intensity of immunoreactive bands specific for human mitochondrial antigen of 60 kDa (mito-P60) relative to total ERK levels in a control cell line, three PINK1shRNA cell lines, and a PINK1-3x-Flag-overexpressing line. The bar graph shows means ± S.E. compiled from 3-4 Western blot experiments for each cell line ( , p < 0.05_versus_ control cell line; n = 5-7 immunoreactive bands analyzed/condition). Inset, representative Western blot showing decreased level of mito-P60 in PINK1 shRNA cell compared with vector stable control (Ctrl), reprobed for total ERK1/2 as a loading control.
, p < 0.05_versus_ control cell line; n = 5-7 immunoreactive bands analyzed/condition). Inset, representative Western blot showing decreased level of mito-P60 in PINK1 shRNA cell compared with vector stable control (Ctrl), reprobed for total ERK1/2 as a loading control.
FIGURE 5.
Increased expression of wild-type PINK1 inhibits autophagy elicited by either 6-OHDA or PINK1 deficiency. A, macroautophagy in PINK1-overexpressing cell lines analyzed by quantifying the average number of GFP-LC3 puncta per cell in the presence of dH20 (vehicle) or 6-OHDA (at 120 μ
m
for 4 h; Sigma) as an inducer of autophagy ( , p < 0.05_versus_ untreated vector control cell line;
, p < 0.05_versus_ untreated vector control cell line; , p < 0.05_versus_ 6-OHDA-treated control cell line (Ctrl); means ± S.E., n = 20-35 cells quantified/condition). B, mitophagy quantification in PINK1-overexpressing clones in the absence or presence of 6-OHDA as an inducer of mitophagy (representative of three experiments;
, p < 0.05_versus_ 6-OHDA-treated control cell line (Ctrl); means ± S.E., n = 20-35 cells quantified/condition). B, mitophagy quantification in PINK1-overexpressing clones in the absence or presence of 6-OHDA as an inducer of mitophagy (representative of three experiments;  , p < 0.05 versus untreated control cell line;
, p < 0.05 versus untreated control cell line; , p < 0.05_versus_ 6-OHDA treated control cell line; 25-30 wells quantified/condition). Restoration of PINK1 expression using the A series RNAi-resistant ΔN-PINK1-3xFLAG plasmid suppresses autophagy in PINK1 shRNA line A14 as assessed by analysis of LC3 shift (C) and in line A4 as assessed by GFP-LC3 puncta (D). (
, p < 0.05_versus_ 6-OHDA treated control cell line; 25-30 wells quantified/condition). Restoration of PINK1 expression using the A series RNAi-resistant ΔN-PINK1-3xFLAG plasmid suppresses autophagy in PINK1 shRNA line A14 as assessed by analysis of LC3 shift (C) and in line A4 as assessed by GFP-LC3 puncta (D). ( , p < 0.0005_versus_ control cell line;
, p < 0.0005_versus_ control cell line; , p < 0.0005_versus_ empty vector transfected PINK1 shRNA; means ± S.E.,n = 15-30 cells quantified/condition.)
, p < 0.0005_versus_ empty vector transfected PINK1 shRNA; means ± S.E.,n = 15-30 cells quantified/condition.)
FIGURE 6.
PINK1 knockdown elicits mitochondrial superoxide upstream of mitochondrial fragmentation and autophagy in PINK1-deficient lines. A, epifluorescence analysis of MitoSOX, a cell-permeable red fluorescent mitochondrial superoxide indicator, in a control stable cell line (Ctrl, left panel) and two PINK1 knockdown cell lines (right panels). Nuclei were counterstained with DRAQ5 (blue). Scale bar: 100 μm. B, bar graph shows compiled means ± S.E. from three independent experiments with increases in the average MitoSOX fluorescence per cell normalized to vector control cell line ( , p < 0.05_versus_ vector control cell line; n = 200-300 cells analyzed/condition). C and D, quantification of mitochondrial interconnectivity (C) and mitochondrial elongation (D) in PINK1 knockdown clonal cell lines treated with catalase, MnTBAP, or vehicle (untreated) for 24 h. (Representative of three independent experiments;
, p < 0.05_versus_ vector control cell line; n = 200-300 cells analyzed/condition). C and D, quantification of mitochondrial interconnectivity (C) and mitochondrial elongation (D) in PINK1 knockdown clonal cell lines treated with catalase, MnTBAP, or vehicle (untreated) for 24 h. (Representative of three independent experiments;  , p < 0.05 versus control cell line; n = 25-35 cells analyzed per condition.) E, representative epifluorescence micrograph of a PINK1 knockdown cell line (A4) transiently expressing GFP-LC3 treated in the presence or absence of antioxidant MnTBAP. Note that MnTBAP suppresses autophagy in PINK1 knockdown cells. F, the average cellular number of GFP-LC3 puncta in control or PINK1shRNA cells transiently expressing GFP-LC3 in the presence or absence of indicated antioxidants. (
, p < 0.05 versus control cell line; n = 25-35 cells analyzed per condition.) E, representative epifluorescence micrograph of a PINK1 knockdown cell line (A4) transiently expressing GFP-LC3 treated in the presence or absence of antioxidant MnTBAP. Note that MnTBAP suppresses autophagy in PINK1 knockdown cells. F, the average cellular number of GFP-LC3 puncta in control or PINK1shRNA cells transiently expressing GFP-LC3 in the presence or absence of indicated antioxidants. ( , p < 0.05_versus_ untreated control cell line;
, p < 0.05_versus_ untreated control cell line; , p < 0.0001_versus_ respective untreated clones; representative bar graph shows means ± S.E., n = 25-40 cells quantified/condition).
, p < 0.0001_versus_ respective untreated clones; representative bar graph shows means ± S.E., n = 25-40 cells quantified/condition).
FIGURE 7.
Mitochondrial changes in PINK1 knockdown cells require Drp1 activity. Control or PINK1shRNA stable clonal cell lines were co-transfected with GFP-Drp1-DN and with a mitochondrially targeted RFP (mito-RFP), and images were analyzed for mitochondrial interconnectivity (A) and mitochondrial elongation (B) as described under “Experimental Procedures.” (Representative of three independent experiments;  , p < 0.005 versus control cell line;
, p < 0.005 versus control cell line; , p < 0.05_versus_ respective empty vector-transfected PINK1shRNA cell line;n = 25-35 cells quantified/condition.) Control or PINK1shRNA stable lines were co-transfected with HA-tagged Drp1-DN and with GFP-LC3 to label AVs. C and D, the average number of GFP-LC3 puncta (C) or the percent colocalization of GFP-LC3 puncta with mitochondria (D) by confocal microscopy and image analysis (representative of three independent experiments;
, p < 0.05_versus_ respective empty vector-transfected PINK1shRNA cell line;n = 25-35 cells quantified/condition.) Control or PINK1shRNA stable lines were co-transfected with HA-tagged Drp1-DN and with GFP-LC3 to label AVs. C and D, the average number of GFP-LC3 puncta (C) or the percent colocalization of GFP-LC3 puncta with mitochondria (D) by confocal microscopy and image analysis (representative of three independent experiments; , p < 0.01_versus_ control;
, p < 0.01_versus_ control;  ,p < 0.05 versus empty vector-transfected A4 or D14;n = 25-30 cells analyzed/condition).
,p < 0.05 versus empty vector-transfected A4 or D14;n = 25-30 cells analyzed/condition).
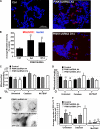
FIGURE 8.
Autophagy is involved in mitochondrial morphology changes elicited by PINK1 deficiency and plays a neuroprotective role. A, stable vector control or PINK1shRNA cells co-transfected with the indicated siRNAs and quantified for the number of GFP-LC3 puncta per cell as analyzed in a representative experiment of two ( , p < 0.001_versus_ control cell line;
, p < 0.001_versus_ control cell line; , p < 0.01_versus_ control siRNA; means ± S.E., n = 30-40 cells quantified per condition). Inset shows a representative Western blot showing effects of Atg7 or control siRNA. Note that knockdown of ATG7 decreases endogenous Atg7 levels and reduces the levels of activated LC3-II.B, the average integrated intensity of MitoSOX fluorescence was quantified in control or PINK1shRNA stable cell lines transiently transfected with the indicated siRNAs (
, p < 0.01_versus_ control siRNA; means ± S.E., n = 30-40 cells quantified per condition). Inset shows a representative Western blot showing effects of Atg7 or control siRNA. Note that knockdown of ATG7 decreases endogenous Atg7 levels and reduces the levels of activated LC3-II.B, the average integrated intensity of MitoSOX fluorescence was quantified in control or PINK1shRNA stable cell lines transiently transfected with the indicated siRNAs ( ,p < 0.01 versus control cell line (Ctrl), means ± S.E., n = 300-400 cells/condition). C and_D_, quantification of mitochondrial interconnectivity (C) and mitochondrial elongation (D) in PINK1 knockdown clonal lines transfected with the indicated siRNAs (representative of three experiments;
,p < 0.01 versus control cell line (Ctrl), means ± S.E., n = 300-400 cells/condition). C and_D_, quantification of mitochondrial interconnectivity (C) and mitochondrial elongation (D) in PINK1 knockdown clonal lines transfected with the indicated siRNAs (representative of three experiments; , p < 0.05_versus_ control cell line transfected with control siRNA;
, p < 0.05_versus_ control cell line transfected with control siRNA; , p < 0.01_versus_ PINK1 A4 or D14 transfected with control siRNA; n = 25-30 cells/condition). E, basal cell survival (Alamar Blue assay) of control and PINK1 shRNA clones (representative of four experiments;
, p < 0.01_versus_ PINK1 A4 or D14 transfected with control siRNA; n = 25-30 cells/condition). E, basal cell survival (Alamar Blue assay) of control and PINK1 shRNA clones (representative of four experiments; , p < 0.005_versus_ control stable cell line; n = 8 wells analyzed/condition). F, propidium iodide (PI) cell death assay of control and PINK1 shRNA stable clonal cell lines (
, p < 0.005_versus_ control stable cell line; n = 8 wells analyzed/condition). F, propidium iodide (PI) cell death assay of control and PINK1 shRNA stable clonal cell lines ( , p < 0.05_versus_ control stable line; n = 800-1000 cells analyzed/condition). G, cell survival of control or PINK1 shRNA stable cell lines treated with bafilomycin or vehicle for 18 h (representative of three experiments;
, p < 0.05_versus_ control stable line; n = 800-1000 cells analyzed/condition). G, cell survival of control or PINK1 shRNA stable cell lines treated with bafilomycin or vehicle for 18 h (representative of three experiments;  ,p < 0.05 versus control cell line/bafilomycin; n = 4 wells analyzed/condition). H, cell survival of control or PINK1shRNA cells transfected with Atg7 siRNA (representative of three experiments;
,p < 0.05 versus control cell line/bafilomycin; n = 4 wells analyzed/condition). H, cell survival of control or PINK1shRNA cells transfected with Atg7 siRNA (representative of three experiments;  , p < 0.05 versus control siRNA-treated clone A4;
, p < 0.05 versus control siRNA-treated clone A4; , p < 0.005_versus_ control siRNA-treated clone D14; n = 6 wells analyzed/condition).
, p < 0.005_versus_ control siRNA-treated clone D14; n = 6 wells analyzed/condition).
FIGURE 9.
Parkin promotes autophagy/mitophagy and restores mitochondria morphology in PINK1 shRNA cell lines. A, representative epifluorescence micrographs of control cells co-transfected with GFP-LC3 and vector (left) and PINK1-deficient cell lines co-transfected with GFP-LC3 and vector (middle) or HA tagged Parkin (right). Cells were fixed and immunostained for HA-tag (red) and counterstained with DAPI to visualize nuclei (blue). B, representative GFP-LC3 puncta bar graph of control cell line and PINK1shRNA cell lines co-transfected with GFP-LC3 and either empty vector or HA-Parkin ( , p < 0.0001_versus_ control cell line;
, p < 0.0001_versus_ control cell line; , p < 0.01_versus_ vector-transfected shRNA cell lines; n = 30-40 cells analyzed per condition). Inset shows representative HA immunoblot for cells transfected with HA tagged Parkin (black arrowhead). The predicted molecular weight of HA-Parkin is 54kDa. C, mitophagy quantification in control and PINK1 shRNA clones transfected with either empty vector or HA-Parkin. (
, p < 0.01_versus_ vector-transfected shRNA cell lines; n = 30-40 cells analyzed per condition). Inset shows representative HA immunoblot for cells transfected with HA tagged Parkin (black arrowhead). The predicted molecular weight of HA-Parkin is 54kDa. C, mitophagy quantification in control and PINK1 shRNA clones transfected with either empty vector or HA-Parkin. ( , p < 0.02 versus control cell line;
, p < 0.02 versus control cell line; , p < 0.05_versus_ PINK1shRNA cell lines; n = 30-40 cells analyzed per condition). D, representative quantification of mitochondrial interconnectivity in a control cell line and PINK1shRNA cell lines co-transfected with mito-GFP and either empty vector or HA-Parkin (
, p < 0.05_versus_ PINK1shRNA cell lines; n = 30-40 cells analyzed per condition). D, representative quantification of mitochondrial interconnectivity in a control cell line and PINK1shRNA cell lines co-transfected with mito-GFP and either empty vector or HA-Parkin ( , p < 0.002_versus_ control cell line;
, p < 0.002_versus_ control cell line; , p < 0.05_versus_PINK1shRNA cell lines; n = 25-30 cells analyzed per condition). E, propidium iodide cell death assay of control and PINK1 shRNA stable clonal cell lines transfected with the indicated plasmids. (
, p < 0.05_versus_PINK1shRNA cell lines; n = 25-30 cells analyzed per condition). E, propidium iodide cell death assay of control and PINK1 shRNA stable clonal cell lines transfected with the indicated plasmids. ( , p < 0.001_versus_ control stable line;
, p < 0.001_versus_ control stable line; , p < 0.05_versus_ PINK1shRNA cell lines; n = 800-1000 cells analyzed/condition).
, p < 0.05_versus_ PINK1shRNA cell lines; n = 800-1000 cells analyzed/condition).
Similar articles
- PGAM5 regulates PINK1/Parkin-mediated mitophagy via DRP1 in CCCP-induced mitochondrial dysfunction.
Park YS, Choi SE, Koh HC. Park YS, et al. Toxicol Lett. 2018 Mar 1;284:120-128. doi: 10.1016/j.toxlet.2017.12.004. Epub 2017 Dec 11. Toxicol Lett. 2018. PMID: 29241732 - Polyphyllin I induces mitophagic and apoptotic cell death in human breast cancer cells by increasing mitochondrial PINK1 levels.
Li GB, Fu RQ, Shen HM, Zhou J, Hu XY, Liu YX, Li YN, Zhang HW, Liu X, Zhang YH, Huang C, Zhang R, Gao N. Li GB, et al. Oncotarget. 2017 Feb 7;8(6):10359-10374. doi: 10.18632/oncotarget.14413. Oncotarget. 2017. PMID: 28060722 Free PMC article. - Parkin and PINK1 functions in oxidative stress and neurodegeneration.
Barodia SK, Creed RB, Goldberg MS. Barodia SK, et al. Brain Res Bull. 2017 Jul;133:51-59. doi: 10.1016/j.brainresbull.2016.12.004. Epub 2016 Dec 23. Brain Res Bull. 2017. PMID: 28017782 Free PMC article. Review. - PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy.
Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N. Kawajiri S, et al. FEBS Lett. 2010 Mar 19;584(6):1073-9. doi: 10.1016/j.febslet.2010.02.016. Epub 2010 Feb 12. FEBS Lett. 2010. PMID: 20153330 - PINK1 and Parkin: team players in stress-induced mitophagy.
Bader V, Winklhofer KF. Bader V, et al. Biol Chem. 2020 May 26;401(6-7):891-899. doi: 10.1515/hsz-2020-0135. Biol Chem. 2020. PMID: 32297878 Review.
Cited by
- The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo.
Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. Vincow ES, et al. Proc Natl Acad Sci U S A. 2013 Apr 16;110(16):6400-5. doi: 10.1073/pnas.1221132110. Epub 2013 Mar 18. Proc Natl Acad Sci U S A. 2013. PMID: 23509287 Free PMC article. - Increased mitochondrial fission promotes autophagy and hepatocellular carcinoma cell survival through the ROS-modulated coordinated regulation of the NFKB and TP53 pathways.
Huang Q, Zhan L, Cao H, Li J, Lyu Y, Guo X, Zhang J, Ji L, Ren T, An J, Liu B, Nie Y, Xing J. Huang Q, et al. Autophagy. 2016 Jun 2;12(6):999-1014. doi: 10.1080/15548627.2016.1166318. Epub 2016 Apr 28. Autophagy. 2016. PMID: 27124102 Free PMC article. - Mitoptosis, a novel mitochondrial death mechanism leading predominantly to activation of autophagy.
Jangamreddy JR, Los MJ. Jangamreddy JR, et al. Hepat Mon. 2012 Aug;12(8):e6159. doi: 10.5812/hepatmon.6159. Epub 2012 Aug 20. Hepat Mon. 2012. PMID: 23087751 Free PMC article. No abstract available. - M-type pyruvate kinase 2 (PKM2) tetramerization alleviates the progression of right ventricle failure by regulating oxidative stress and mitochondrial dynamics.
Guo L, Wang L, Qin G, Zhang J, Peng J, Li L, Chen X, Wang D, Qiu J, Wang E. Guo L, et al. J Transl Med. 2023 Dec 7;21(1):888. doi: 10.1186/s12967-023-04780-6. J Transl Med. 2023. PMID: 38062516 Free PMC article. - PINK1: A Bridge between Mitochondria and Parkinson's Disease.
Gonçalves FB, Morais VA. Gonçalves FB, et al. Life (Basel). 2021 Apr 21;11(5):371. doi: 10.3390/life11050371. Life (Basel). 2021. PMID: 33919398 Free PMC article. Review.
References
- Hoepken, H. H., Gispert, S., Morales, B., Wingerter, O., Del Turco, D., Mulsch, A., Nussbaum, R. L., Muller, K., Drose, S., Brandt, U., Deller, T., Wirth, B., Kudin, A. P., Kunz, W. S., and Auburger, G. (2007) Neurobiol. Dis. 25 401-411 - PubMed
- Exner, N., Treske, B., Paquet, D., Holmstrom, K., Schiesling, C., Gispert, S., Carballo-Carbajal, I., Berg, D., Hoepken, H. H., Gasser, T., Kruger, R., Winklhofer, K. F., Vogel, F., Reichert, A. S., Auburger, G., Kahle, P. J., Schmid, B., and Haass, C. (2007) J. Neurosci. 27 12413-12418 - PMC - PubMed
- Abeliovich, A., and Flint Beal, M. (2006) J. Neurochem. 99 1062-1072 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS05377/NS/NINDS NIH HHS/United States
- K18 DC009120-01/DC/NIDCD NIH HHS/United States
- K18 DC009120/DC/NIDCD NIH HHS/United States
- F32 AG030821/AG/NIA NIH HHS/United States
- R21 NS053777-01/NS/NINDS NIH HHS/United States
- R01 AG026389/AG/NIA NIH HHS/United States
- R01 AG026389-02/AG/NIA NIH HHS/United States
- R21 NS053777/NS/NINDS NIH HHS/United States
- AG026389/AG/NIA NIH HHS/United States
- R01 AG026389-01A2/AG/NIA NIH HHS/United States
- DC009120/DC/NIDCD NIH HHS/United States
- R21 NS053777-02/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous