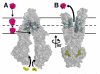Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding - PubMed (original) (raw)
Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding
Stephen G Aller et al. Science. 2009.
Abstract
P-glycoprotein (P-gp) detoxifies cells by exporting hundreds of chemically unrelated toxins but has been implicated in multidrug resistance (MDR) in the treatment of cancers. Substrate promiscuity is a hallmark of P-gp activity, thus a structural description of poly-specific drug-binding is important for the rational design of anticancer drugs and MDR inhibitors. The x-ray structure of apo P-gp at 3.8 angstroms reveals an internal cavity of approximately 6000 angstroms cubed with a 30 angstrom separation of the two nucleotide-binding domains. Two additional P-gp structures with cyclic peptide inhibitors demonstrate distinct drug-binding sites in the internal cavity capable of stereoselectivity that is based on hydrophobic and aromatic interactions. Apo and drug-bound P-gp structures have portals open to the cytoplasm and the inner leaflet of the lipid bilayer for drug entry. The inward-facing conformation represents an initial stage of the transport cycle that is competent for drug binding.
Figures
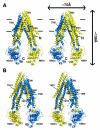
Fig. 1
Structure of Pgp. (A) Front and (B) back stereo views of PGP. TM1-12 are labeled. The N- and C-terminal half of the molecule is colored yellow and blue, respectively. TM4-5 and TM10-11 cross over to form intertwined interfaces that stabilize the inward facing conformation. Horizontal bars represent the approximate positioning of the lipid bilayer. The N- and C-termini are labeled in panel A. Transmembrane (TM) domains and nucleotide binding domains (NBD) are also labeled.
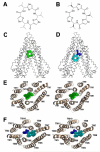
Fig. 2
Binding of novel cyclic peptide Pgp inhibitors. Chemical structures of (A) QZ59-RRR and (B) QZ59-SSS. (C) Location of one QZ59-RRR (green spheres) and (D) two QZ59-SSS (blue and cyan spheres) molecules in the Pgp internal cavity. (E-F) Stereo images showing interaction of transmembrane helices with QZ59 compounds viewed from the intracellular side of the protein looking into the internal chamber. In both cases the compound(s) are sandwiched between previously identified drug binding TMs 6 and 12. The location of the QZ59 compounds was verified by anomalous Fourier (Fig. S15B-C) and Fo-Fc maps (Fig. S15D-E and S16-S17).
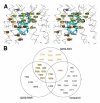
Fig. 3
Drug binding residues of Pgp. (A) Stereo view of the drug-binding cavity. Cα trace shown in gray. The QZ59-SSS in the “lower” (cyan) and “upper” (blue), as well as QZ59-RRR occupying the “middle” site (green) are superimposed. Residues within ∼4-5 Å of QZ59 compounds are shown as spheres. Spheres colored orange and red represent residues that only contact QZ59-SSS in the “lower” and “upper” site, respectively. Residues in common between QZ59-RRR and QZ59-SSS sites are colored yellow. Four residues (grey spheres) are close to QZ59-RRR but neither QZ59-SSS molecules. (B) Venn diagram of residues in close proximity to QZ59 molecules and residues that are protected from MTS labeling by verapamil binding (Fig. S1) (20, 21). Only residues in contact with the model for QZ59-SSS are displayed. Verapamil-only interacting residues are omitted from (A) for clarity and shown in Fig. S16.
Fig. 4
Model of substrate transport by Pgp. (A) Substrate (magenta) partitions into the bilayer from outside of the cell to the inner leaflet and enters the internal drug-binding pocket through an open portal. The residues in the drug binding pocket (cyan spheres) interact with QZ59 compounds and verapamil in the inward facing conformation. (B) ATP (yellow) binds to the NBDs causing a large conformational change presenting the substrate and drug-binding site(s) to the outer leaflet/extracellular space. In this model of Pgp, which is based on the outward facing conformation of MsbA and Sav1866 (13, 14), exit of the substrate to the inner leaflet is sterically occluded providing unidirectional transport to the outside.
Comment in
- Biochemistry. Through a mirror, differently.
Sheps JA. Sheps JA. Science. 2009 Mar 27;323(5922):1679-80. doi: 10.1126/science.1172428. Science. 2009. PMID: 19325102 No abstract available.
Similar articles
- Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans.
Jin MS, Oldham ML, Zhang Q, Chen J. Jin MS, et al. Nature. 2012 Oct 25;490(7421):566-9. doi: 10.1038/nature11448. Epub 2012 Sep 23. Nature. 2012. PMID: 23000902 Free PMC article. - The importance of cholesterol in maintenance of P-glycoprotein activity and its membrane perturbing influence.
Rothnie A, Theron D, Soceneantu L, Martin C, Traikia M, Berridge G, Higgins CF, Devaux PF, Callaghan R. Rothnie A, et al. Eur Biophys J. 2001 Oct;30(6):430-42. doi: 10.1007/s002490100156. Eur Biophys J. 2001. PMID: 11718296 - Characterization of two pharmacophores on the multidrug transporter P-glycoprotein.
Garrigues A, Loiseau N, Delaforge M, Ferté J, Garrigos M, André F, Orlowski S. Garrigues A, et al. Mol Pharmacol. 2002 Dec;62(6):1288-98. doi: 10.1124/mol.62.6.1288. Mol Pharmacol. 2002. PMID: 12435795 - P-glycoprotein--implications of metabolism of neoplastic cells and cancer therapy.
Breier A, Barancík M, Sulová Z, Uhrík B. Breier A, et al. Curr Cancer Drug Targets. 2005 Sep;5(6):457-68. doi: 10.2174/1568009054863636. Curr Cancer Drug Targets. 2005. PMID: 16178819 Review. - Structural and functional aspects of P-glycoprotein and its inhibitors.
Mollazadeh S, Sahebkar A, Hadizadeh F, Behravan J, Arabzadeh S. Mollazadeh S, et al. Life Sci. 2018 Dec 1;214:118-123. doi: 10.1016/j.lfs.2018.10.048. Epub 2018 Oct 27. Life Sci. 2018. PMID: 30449449 Review.
Cited by
- Gastrointestinal Bleeding Due to NOACs Use: Exploring the Molecular Mechanisms.
Saviano A, Brigida M, Petruzziello C, Candelli M, Gabrielli M, Ojetti V. Saviano A, et al. Int J Mol Sci. 2022 Nov 12;23(22):13955. doi: 10.3390/ijms232213955. Int J Mol Sci. 2022. PMID: 36430433 Free PMC article. Review. - Phosphorylation-dependent changes in nucleotide binding, conformation, and dynamics of the first nucleotide binding domain (NBD1) of the sulfonylurea receptor 2B (SUR2B).
de Araujo ED, Alvarez CP, López-Alonso JP, Sooklal CR, Stagljar M, Kanelis V. de Araujo ED, et al. J Biol Chem. 2015 Sep 11;290(37):22699-714. doi: 10.1074/jbc.M114.636233. Epub 2015 Jul 21. J Biol Chem. 2015. PMID: 26198630 Free PMC article. - Advances in PET imaging of P-glycoprotein function at the blood-brain barrier.
Syvänen S, Eriksson J. Syvänen S, et al. ACS Chem Neurosci. 2013 Feb 20;4(2):225-37. doi: 10.1021/cn3001729. Epub 2012 Dec 4. ACS Chem Neurosci. 2013. PMID: 23421673 Free PMC article. Review. - Inhibition of P-glycoprotein Gene Expression and Function Enhances Triptolide-induced Hepatotoxicity in Mice.
Kong LL, Zhuang XM, Yang HY, Yuan M, Xu L, Li H. Kong LL, et al. Sci Rep. 2015 Jul 2;5:11747. doi: 10.1038/srep11747. Sci Rep. 2015. PMID: 26134275 Free PMC article. - Determining P-glycoprotein-drug interactions: evaluation of reconstituted P-glycoprotein in a liposomal system and LLC-MDR1 polarized cell monolayers.
Melchior DL, Sharom FJ, Evers R, Wright GE, Chu JW, Wright SE, Chu X, Yabut J. Melchior DL, et al. J Pharmacol Toxicol Methods. 2012 Mar;65(2):64-74. doi: 10.1016/j.vascn.2012.02.002. Epub 2012 Feb 26. J Pharmacol Toxicol Methods. 2012. PMID: 22394995 Free PMC article.
References
- The American Cancer Society Homepage Global Cancer Facts and Figures. 2007. [Accessed Sept. 4, 2008]. http://www.cancer.org/downloads/STT/Global_Cancer_Facts_and_Figures_2007....
- Longley DB, Johnston PG. Journal of Pathology. 2005 Jan;205:275. - PubMed
- Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Nature Reviews Drug Discovery. 2006 Mar;5:219. - PubMed
- Sharom FJ. Pharmacogenomics. 2008 Jan;9:105. - PubMed
- Ramachandra M, et al. Biochemistry. 1998 Apr 7;37:5010. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM078914-03/GM/NIGMS NIH HHS/United States
- GM078914/GM/NIGMS NIH HHS/United States
- P50 GM073197/GM/NIGMS NIH HHS/United States
- GM61905/GM/NIGMS NIH HHS/United States
- R01 GM061905-09/GM/NIGMS NIH HHS/United States
- R01 GM061905/GM/NIGMS NIH HHS/United States
- F32 GM078914/GM/NIGMS NIH HHS/United States
- P50 GM073197-050002/GM/NIGMS NIH HHS/United States
- GM073197/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous