Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades - PubMed (original) (raw)
Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades
Shani Blanga-Kanfi et al. BMC Evol Biol. 2009.
Abstract
Background: Rodentia is the most diverse order of placental mammals, with extant rodent species representing about half of all placental diversity. In spite of many morphological and molecular studies, the family-level relationships among rodents and the location of the rodent root are still debated. Although various datasets have already been analyzed to solve rodent phylogeny at the family level, these are difficult to combine because they involve different taxa and genes.
Results: We present here the largest protein-coding dataset used to study rodent relationships. It comprises six nuclear genes, 41 rodent species, and eight outgroups. Our phylogenetic reconstructions strongly support the division of Rodentia into three clades: (1) a "squirrel-related clade", (2) a "mouse-related clade", and (3) Ctenohystrica. Almost all evolutionary relationships within these clades are also highly supported. The primary remaining uncertainty is the position of the root. The application of various models and techniques aimed to remove non-phylogenetic signal was unable to solve the basal rodent trifurcation.
Conclusion: Sequencing and analyzing a large sequence dataset enabled us to resolve most of the evolutionary relationships among Rodentia. Our findings suggest that the uncertainty regarding the position of the rodent root reflects the rapid rodent radiation that occurred in the Paleocene rather than the presence of conflicting phylogenetic and non-phylogenetic signals in the dataset.
Figures
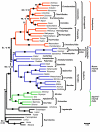
Figure 1
ML tree (-ln likelihood = 85,018.88) obtained for the concatenated dataset under the GTR+Γ+I model of sequence evolution. For each node the ML bootstrap percentage (BP) and the Bayesian posterior probabilities (PP) are given at the right and left of the slash, respectively. Branches with maximal support value (i.e., BP = 100, PP = 1.0) are indicated by black dots.
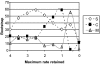
Figure 2
Bootstrap support as a function of the maximum evolutionary rate of site retained in the data. S: the best topology with the squirrel-related clade at the base of the rodent tree; M: the best topology with the mouse-related clade at the base of the rodent tree; C: the best topology with Ctenohystrica at the base of the rodent tree. Nine datasets were considered: (1) all sites (6,255 base pairs (bps); rates range from -0.698 to 3.989); (2) sites with rate ≤ 3.5 (6,114 bps); (3) sites with rate ≤ 3.0 (6,058 bps); (4) sites with rate ≤ 2.5 (5,997 bps); (5) sites with rate ≤ 2.0 (5,896 bps); (6) sites with rate ≤ 1.5 (5,759 bps); (7) sites with rate ≤ 1 (5,444 bps); (8) sites with rate ≤ 0.5 (4,997 bps); and (9) sites with rate ≤ 0.0 (4,179 bps).
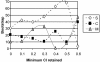
Figure 3
Bootstrap support as a function of the minimum CI of sites retained in the data. S: the best topology with the squirrel-related clade at the base of the rodent tree; M: the best topology with the mouse-related clade at the base of the rodent tree; C: the best topology with Ctenohystrica at the base of the rodent tree. Seven datasets were considered: (1) all sites (6,255 bps; CI range 0.0625–1); (2) sites with CI > 0.1 (6,210 bps); (3) sites with CI > 0.2 (5,864 bps); (4) sites with CI > 0.3 (5,432 bps); (5) sites with CI > 0.4 (4,833 bps); (6) sites with CI > 0.5 (4,119 bps); and (7) sites with CI > 0.6 (4,023 bps).
Similar articles
- A glimpse on the pattern of rodent diversification: a phylogenetic approach.
Fabre PH, Hautier L, Dimitrov D, Douzery EJ. Fabre PH, et al. BMC Evol Biol. 2012 Jun 14;12:88. doi: 10.1186/1471-2148-12-88. BMC Evol Biol. 2012. PMID: 22697210 Free PMC article. - Suprafamilial relationships among Rodentia and the phylogenetic effect of removing fast-evolving nucleotides in mitochondrial, exon and intron fragments.
Montgelard C, Forty E, Arnal V, Matthee CA. Montgelard C, et al. BMC Evol Biol. 2008 Nov 26;8:321. doi: 10.1186/1471-2148-8-321. BMC Evol Biol. 2008. PMID: 19036132 Free PMC article. - Rodent phylogeny and a timescale for the evolution of Glires: evidence from an extensive taxon sampling using three nuclear genes.
Huchon D, Madsen O, Sibbald MJ, Ament K, Stanhope MJ, Catzeflis F, de Jong WW, Douzery EJ. Huchon D, et al. Mol Biol Evol. 2002 Jul;19(7):1053-65. doi: 10.1093/oxfordjournals.molbev.a004164. Mol Biol Evol. 2002. PMID: 12082125 - Phylogeny of rodentia (Mammalia) inferred from the nuclear-encoded gene IRBP.
DeBry RW, Sagel RM. DeBry RW, et al. Mol Phylogenet Evol. 2001 May;19(2):290-301. doi: 10.1006/mpev.2001.0945. Mol Phylogenet Evol. 2001. PMID: 11341810
Cited by
- Springing into action: Comparing escape responses between bipedal and quadrupedal rodents.
Freymiller GA, Whitford MD, McGowan CP, Higham TE, Clark RW. Freymiller GA, et al. Ecol Evol. 2024 Sep 20;14(9):e70292. doi: 10.1002/ece3.70292. eCollection 2024 Sep. Ecol Evol. 2024. PMID: 39310732 Free PMC article. - Epithelial Na + Channels Function as Extracellular Sensors.
Kashlan OB, Wang XP, Sheng S, Kleyman TR. Kashlan OB, et al. Compr Physiol. 2024 Mar 29;14(2):1-41. doi: 10.1002/cphy.c230015. Compr Physiol. 2024. PMID: 39109974 Review. - Evolutionary Patterns of Maternal Recognition of Pregnancy and Implantation in Eutherian Mammals.
Braz HB, Barreto RDSN, Silva-Júnior LND, Horvath-Pereira BO, Silva TSD, Silva MDD, Acuña F, Miglino MA. Braz HB, et al. Animals (Basel). 2024 Jul 16;14(14):2077. doi: 10.3390/ani14142077. Animals (Basel). 2024. PMID: 39061539 Free PMC article. Review. - Shrinkage-based Random Local Clocks with Scalable Inference.
Fisher AA, Ji X, Nishimura A, Baele G, Lemey P, Suchard MA. Fisher AA, et al. Mol Biol Evol. 2023 Nov 3;40(11):msad242. doi: 10.1093/molbev/msad242. Mol Biol Evol. 2023. PMID: 37950885 Free PMC article.
References
- Wilson DE, Reeder DM. Mammal species of the world: a taxonomic and geographic reference. 3. Baltimore, MD, Johns Hopkins University Press; 2005.
- Luckett WP, Hartenberger J-L. Monophyly or polyphyly of the order Rodentia: possible conflict between morphological and molecular interpretations. J Mammal Evol. 1993;1:127–147. doi: 10.1007/BF01041591. - DOI
- Hartenberger J-L. The order Rodentia: major questions on their evolutionary origin, relationships and suprafamilial systematics. In: Luckett WP, Hartenberger J-L, editor. Evolutionary relationships among rodents: a multidisciplinary analysis. New York and London, Plenum Press; 1985. pp. 1–33.
- McKenna MC, Bell SK. Classification of mammals above the species level. New York, Columbia University Press; 1997.
- Jaeger J-J. Rodent phylogeny: new data and old problems. In: Benton MJ, editor. The phylogeny and classification of the Tetrapods. Vol. 2. Oxford, Clarendon Press; 1988. pp. 177–199.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources