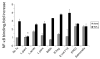Bacteria-free solution derived from Lactobacillus plantarum inhibits multiple NF-kappaB pathways and inhibits proteasome function - PubMed (original) (raw)
Bacteria-free solution derived from Lactobacillus plantarum inhibits multiple NF-kappaB pathways and inhibits proteasome function
Elaine O Petrof et al. Inflamm Bowel Dis. 2009 Oct.
Abstract
Background: Bacteria play a role in inflammatory bowel disease and other forms of intestinal inflammation. Although much attention has focused on the search for a pathogen or inciting inflammatory bacteria, another possibility is a lack of beneficial bacteria that normally confer anti-inflammatory properties in the gut. The purpose of this study was to determine whether normal commensal bacteria could inhibit inflammatory pathways important in intestinal inflammation.
Methods: Conditioned media from Lactobacillus plantarum (Lp-CM) and other gut bacteria was used to treat intestinal epithelial cell (YAMC) and macrophage (RAW 264.7) or primary culture murine dendritic cells. NF-kappaB was activated through TNF-Receptor, MyD88-dependent and -independent pathways and effects of Lp-CM on the NF-kappaB pathway were assessed. NF-kappaB binding activity was measured using ELISA and EMSA. 1kappaB expression was assessed by Western blot analysis, and proteasome activity determined using fluorescence-based proteasome assays. MCP-1 release was determined by ELISA.
Results: Lp-CM inhibited NF-kappaB binding activity, degradation of IkappaBalpha and the chymotrypsin-like activity of the proteasome. Moreover, Lp-CM directly inhibited the activity of purified mouse proteasomes. This effect was specific, since conditioned media from other bacteria had no inhibitory effect. Unlike other proteasome inhibitors, Lp-CM was not toxic in cell death assays. Lp-CM inhibited MCP-1 release in all cell types tested.
Conclusions: These studies confirm, and provide a mechanism for, the anti-inflammatory effects of the probiotic and commensal bacterium Lactobacillus plantarum. The use of bacteria-free Lp-CM provides a novel strategy for treatment of intestinal inflammation which would eliminate the risk of bacteremia reported with conventional probiotics.
Figures
FIGURE 1
Assessment of intestinal bacteria conditioned media to inhibit NF-_κ_B binding in gut epithelial cells. YAMC cells were either pretreated with conditioned media alone from the intestinal bacteria indicated (gray bars), or pre-treated and then stimulated with 30 ng/mL TNF-α for 30 minutes (black bars). A commercially available NF-_κ_B ELISA (Active Motif) was then used to test nuclear extracts and determine degree of NF-_κ_B binding. Results are expressed as mean ± SD for a minimum of 3 separate experiments (*P < 0.01 by ANOVA analysis using Bonferroni correction).
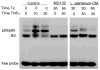
FIGURE 2
Lp-CM inhibits NF-_κ_B binding in intestinal epithelial cells. YAMC cells were pretreated with either MG132 or Lp-CM for the times indicated (Time Tx), and then treated with 30 ng/mL TNF-α for the times indicated prior to nuclear extract harvest and electrophoretic mobility shift assay (EMSA). Pretreatment with Lp-CM blocks TNF-α-induced NF-_κ_B binding. Nonspecific binding (NS) indicated, free probe is at bottom of gel. Blot shown is representative of 3 independent experiments.
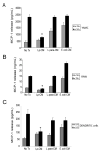
FIGURE 3
Lp-CM inhibits TNF-α-mediated MCP-1 release in multiple cell types. A: YAMC cells were treated with either Lp-CM, _L. paracasei_-CM, or _E. coli_-CM for 4 hours, and then with TNF-α (30 ng/mL) to stimulate NF-_κ_B. Supernatants were harvested after 6 hours and and then assayed for release of the chemokine MCP-1, a downstream gene target of NF-_κ_B, using a commercially available ELISA. Results were compared to vehicle-treated control (No Treatment, column 1), or TNF-α treatment alone (column 2). Results are expressed as mean ± SD for 3 separate experiments (*P < 0.01 by ANOVA analysis using Bonferroni correction). B: RAW 264.7 murine macrophage cells were treated with either Lp-CM, _L. paracasei_-CM, or _E. coli_-CM for 4 hours, then with TNF-α (30 ng/mL) to activate NF-_κ_B. Supernatants were harvested after 6 hours and and then assayed for release of MCP-1 using a commercially available ELISA. Results are expressed as mean ± SD for 3 separate experiments (*P < 0.01 by ANOVA analysis using Bonferroni correction). C: Primary murine dendritic cells were prepared as described in Materials and Methods and then treated with either Lp-CM, _L. paracasei_-CM, or _E. coli_-CM for 3 hours, followed by LPS treatment for 16 hours to activate NF-_κ_B. Results are expressed as mean ± SD for 3 separate experiments (*P < 0.01 by ANOVA analysis using Bonferroni correction).
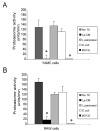
FIGURE 4
Lp-CM inhibits proteasome activity in different cell types. A: YAMC cells were treated with Lp-CM, _L. paracasei_-CM, or _E. coli_-CM or the synthetic proteasome inhibitor MG132 for 1 hour, then harvested for proteasome assay using SLLVY-AMC substrate (see text). Results are expressed as proteasome activity in fluorescence units/min over time. Activity was determined by measuring fluorescence (excitation 380 nm, emission 460 nm) every 3 minutes for 30 minutes to determine reaction rate. Results are expressed as mean ± SD for 3 separate experiments (*P < 0.01 by ANOVA with Bonferroni correction). B: RAW 264.7 macrophage cells were treated with Lp-CM, _L. paracasei_-CM, or _E. coli_-CM or the synthetic proteasome inhibitor MG132 for 1 hour, then harvested for proteasome assay using SLLVY-AMC substrate as in panel A. Results are expressed as mean ± SD for 3 separate experiments (*P < 0.01 by ANOVA with Bonferroni correction).
FIGURE 5
I_κ_B_α_ degradation is inhibited by Lp-CM. YAMC cells were pretreated with either vehicle control (HBSS) or with Lp-CM for 1 hour, then stimulated with TNF-α (30 ng/mL) and harvested at the timepoints (min) indicated. Samples were then subjected to Western blot analysis for the presence of I_κ_Bα. Hsc73 (heat shock cognate 73) was used as a loading control. Blot shown is representative of 3 separate experiments.
FIGURE 6
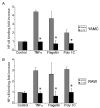
Pretreatment with Lp-CM inhibits NF-_κ_B activation by multiple signaling pathways. Lp-CM was used to treat YAMC cells, an intestinal epithelial cell line derived from mouse large intestine (A) or RAW 264.7 cells, a murine macrophage cell line (B). Cells were activated either with 100 ng/mL flagellin (TLR5 ligand, MyD88-dependent), or with poly dI:dC (TLR3 ligand, MyD88-independent) and NF-_κ_B activity was tested using an NF-_κ_B ELISA assay (Active Motif) as described in Materials and Methods. TNF-α activation is also shown. Data are presented as mean ± SD for 3 separate experiments and vehicle (gray bars) versus Lp-CM treated sample (black bars) pairs were compared using Student’s _t_-test (P < 0.05 was accepted as a level of statistically significant difference).
FIGURE 7
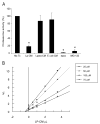
Lp-CM directly inhibits proteasome activity and is a reversible proteasome inhibitor. A: Proteasomes were purified from mouse liver as described in Materials and Methods, treated with Lp-CM, _L. paracasei_-CM, or _E. coli_-CM, and then proteasome assays were performed immediately to determine whether the effect of Lp-CM was a direct inhibitory effect on the proteasome. A fluorometer was used to record measurements every minute for 30 minutes (excitation 380 nm, emission 460 nm) and slopes were calculated using CaryEclipse and KC4 software (see Materials and Methods). Results are expressed as percent activity, with untreated proteasome preparations defined as 100% activity (y-axis). Epoxomicin, a highly specific proteasome inhibitor, was used as an inhibitor control to ensure absence of other contaminating proteases. MG132 is also shown. Results are expressed as mean ± SD for 3 separate experiments (*P < 0.01 by ANOVA with Bonferroni correction). B: Proteasome assays were performed using varying concentrations of SLLVY-AMC substrate and varying concentrations of Lp-CM as indicated. Reaction rates were used within the first 5 minutes of starting the assay in order to determine Vi (initial reaction rate) for generation of the Dixon plot data. Dixon plots were generated using Grafit software.
FIGURE 8
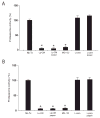
Bioactive factors in Lp-CM are heat-stable and protease-resistant. A: Purified proteasomes from mouse liver were treated with Lp-CM or L. paracasei-CM that was thoroughly boiled for 10 minutes. Treatments were added directly to the proteasome preparation, SLLVY-AMC substrate was added, and the samples assayed for proteasome activity as previously described (excitation 380 nm, emission 460 nm). Slopes were calculated using CaryEclipse software (see Materials and Methods) and results expressed as percent activity, with untreated proteasome preparations defined as 100% activity (y-axis). MG132 is also shown. Results are expressed as mean ± SD for 3 separate experiments (*P < 0.01 by ANOVA with Bonferroni correction). B: Proteasome assays were performed as in panel A except that Lp-CM and _L. paracasei_-CM were subjected to protease digestion using a pepsin digestion method as previously described. Results are expressed as mean ± SD for 3 separate experiments (*P < 0.01 by ANOVA with Bonferroni correction).
FIGURE 9
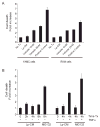
Lp-CM does not induce increased cell death compared to other proteasome inhibitors. A: Lp-CM was used to treat YAMC cells or RAW cells for 4 hours and the effects of Lp-CM on cell death were determined using a commercially available cell death ELISA assay. The proteasome inhibitor bortezomib is also shown. Results are expressed as mean ± SD for 3 separate experiments. B: YAMC cells were pretreated with either MG132 or Lp-CM for the times indicated (0--8 hours). Cells pretreated with either MG132 or Lp-CM for 4 hours were then treated with 30 ng/mL TNF-α for an additional 4 hours and cell death measured by ELISA. Results are expressed as mean ± SD for 2 separate experiments, performed in triplicate.
Similar articles
- Synergistic protection of combined probiotic conditioned media against neonatal necrotizing enterocolitis-like intestinal injury.
Shiou SR, Yu Y, Guo Y, He SM, Mziray-Andrew CH, Hoenig J, Sun J, Petrof EO, Claud EC. Shiou SR, et al. PLoS One. 2013 May 24;8(5):e65108. doi: 10.1371/journal.pone.0065108. Print 2013. PLoS One. 2013. PMID: 23717690 Free PMC article. - Probiotics inhibit nuclear factor-kappaB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition.
Petrof EO, Kojima K, Ropeleski MJ, Musch MW, Tao Y, De Simone C, Chang EB. Petrof EO, et al. Gastroenterology. 2004 Nov;127(5):1474-87. doi: 10.1053/j.gastro.2004.09.001. Gastroenterology. 2004. PMID: 15521016 - Suppression of NF-kappaB-dependent proinflammatory gene expression in human RPE cells by a proteasome inhibitor.
Wang XC, Jobin C, Allen JB, Roberts WL, Jaffe GJ. Wang XC, et al. Invest Ophthalmol Vis Sci. 1999 Feb;40(2):477-86. Invest Ophthalmol Vis Sci. 1999. PMID: 9950608 - Interplay between proteasome inhibitors and NF-κB pathway in leukemia and lymphoma: a comprehensive review on challenges ahead of proteasome inhibitors.
Pakjoo M, Ahmadi SE, Zahedi M, Jaafari N, Khademi R, Amini A, Safa M. Pakjoo M, et al. Cell Commun Signal. 2024 Feb 8;22(1):105. doi: 10.1186/s12964-023-01433-5. Cell Commun Signal. 2024. PMID: 38331801 Free PMC article. Review. - Basic and clinical research on the regulation of the intestinal barrier by Lactobacillus and its active protein components: a review with experience of one center.
Liu ZH, Kang L, Wang JP. Liu ZH, et al. Mol Biol Rep. 2014 Dec;41(12):8037-46. doi: 10.1007/s11033-014-3701-9. Epub 2014 Sep 4. Mol Biol Rep. 2014. PMID: 25185994 Review.
Cited by
- Novel perspectives in probiotic treatment: the efficacy and unveiled mechanisms of the physiological functions.
Fujiya M, Kohgo Y. Fujiya M, et al. Clin J Gastroenterol. 2010 Jun;3(3):117-27. doi: 10.1007/s12328-010-0154-0. Epub 2010 May 8. Clin J Gastroenterol. 2010. PMID: 26190117 - Potential anti-ageing effects of probiotic-derived conditioned media on human skin cells.
Hong YK, An S, Lee YH, Yang SA, Yoon YK, Lee J, Lee G, Chung MJ, Bae S. Hong YK, et al. Acta Pharm. 2022 Apr 13;72(3):359-374. doi: 10.2478/acph-2022-0027. Print 2022 Sep 1. Acta Pharm. 2022. PMID: 36651546 - Early mucosal sensing of SIV infection by paneth cells induces IL-1β production and initiates gut epithelial disruption.
Hirao LA, Grishina I, Bourry O, Hu WK, Somrit M, Sankaran-Walters S, Gaulke CA, Fenton AN, Li JA, Crawford RW, Chuang F, Tarara R, Marco ML, Bäumler AJ, Cheng H, Dandekar S. Hirao LA, et al. PLoS Pathog. 2014 Aug 28;10(8):e1004311. doi: 10.1371/journal.ppat.1004311. eCollection 2014 Aug. PLoS Pathog. 2014. PMID: 25166758 Free PMC article. - Classical and recent advances in the treatment of inflammatory bowel diseases.
Sales-Campos H, Basso PJ, Alves VB, Fonseca MT, Bonfá G, Nardini V, Cardoso CR. Sales-Campos H, et al. Braz J Med Biol Res. 2015 Feb;48(2):96-107. doi: 10.1590/1414-431X20143774. Epub 2014 Nov 28. Braz J Med Biol Res. 2015. PMID: 25466162 Free PMC article. Review. - Probiotics: progress toward novel therapies for intestinal diseases.
Yan F, Polk DB. Yan F, et al. Curr Opin Gastroenterol. 2010 Mar;26(2):95-101. doi: 10.1097/MOG.0b013e328335239a. Curr Opin Gastroenterol. 2010. PMID: 19952741 Free PMC article. Review.
References
- Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. - PubMed
- Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. - PubMed
- Yan F, Polk DB. Probiotics as functional food in the treatment of diarrhea. Curr Opin Clin Nutr Metab Care. 2006;9:717–721. - PubMed
- Kassinen A, Krogius-Kurikka L, Makivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 HD043839-06/HD/NICHD NIH HHS/United States
- P30 DK042086/DK/NIDDK NIH HHS/United States
- K08 DK064840-01/DK/NIDDK NIH HHS/United States
- P30 DK042086-199009/DK/NIDDK NIH HHS/United States
- K08 DK064840-04/DK/NIDDK NIH HHS/United States
- DK064840/DK/NIDDK NIH HHS/United States
- DK42086/DK/NIDDK NIH HHS/United States
- R37 DK047722/DK/NIDDK NIH HHS/United States
- R37 DK047722-14A1/DK/NIDDK NIH HHS/United States
- P30 DK042086-199008/DK/NIDDK NIH HHS/United States
- K08 HD043839/HD/NICHD NIH HHS/United States
- DK47722/DK/NIDDK NIH HHS/United States
- AT004044/AT/NCCIH NIH HHS/United States
- P30 DK042086-199010/DK/NIDDK NIH HHS/United States
- K08 DK064840-03/DK/NIDDK NIH HHS/United States
- P30 DK042086-199011/DK/NIDDK NIH HHS/United States
- R01 DK047722-08/DK/NIDDK NIH HHS/United States
- HD043839/HD/NICHD NIH HHS/United States
- R21 AT004044-01A2/AT/NCCIH NIH HHS/United States
- R21 AT004044/AT/NCCIH NIH HHS/United States
- K08 DK064840/DK/NIDDK NIH HHS/United States
- K08 DK064840-05/DK/NIDDK NIH HHS/United States
- R01 DK047722/DK/NIDDK NIH HHS/United States
- K08 DK064840-02/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous