Composition and three-dimensional architecture of the dengue virus replication and assembly sites - PubMed (original) (raw)
Composition and three-dimensional architecture of the dengue virus replication and assembly sites
Sonja Welsch et al. Cell Host Microbe. 2009.
Abstract
Positive-strand RNA viruses are known to rearrange cellular membranes to facilitate viral genome replication. The biogenesis and three-dimensional organization of these membranes and the link between replication and virus assembly sites is not fully clear. Using electron microscopy, we find Dengue virus (DENV)-induced vesicles, convoluted membranes, and virus particles to be endoplasmic reticulum (ER)-derived, and we detect double-stranded RNA, a presumed marker of RNA replication, inside virus-induced vesicles. Electron tomography (ET) shows DENV-induced membrane structures to be part of one ER-derived network. Furthermore, ET reveals vesicle pores that could enable release of newly synthesized viral RNA and reveals budding of DENV particles on ER membranes directly apposed to vesicle pores. Thus, DENV modifies ER membrane structure to promote replication and efficient encapsidation of the genome into progeny virus. This architecture of DENV replication and assembly sites could explain the coordination of distinct steps of the flavivirus replication cycle.
Figures
Figure 1
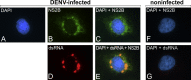
Colocalization of DENV NS2B with Double-Stranded RNA in Infected Cells (A–G) Huh-7 cells infected with DENV2 (A–E) or none-infected (F and G) were fixed 24 hr postinfection and labeled with a monospecific polyclonal rabbit antiserum to DENV NS2B and a monospecific mouse monoclonal antibody against dsRNA. Nuclear DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride).
Figure 2
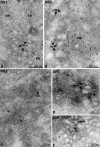
Ultrastructure of DENV-Induced Membrane Alterations (A) Thin-section TEM images of DENV-infected, resin-embedded Huh-7 cells fixed at 24 hr postinfection are shown in (A)–(D). Overview of the virus-induced structures is shown in (A); virus-induced vesicles (Ve) and occasionally membrane tubes (T) accumulate around stacks of convoluted membranes (CMs). Ve and CM are intimately associated with the ER. Clusters of virus particles (Vi) reside in the periphery of virus-induced membrane structures and are aligned in stacks in the lumen of dilated ER cisternae. (B) Arrays of CM and associated Ve, surrounded by ER membranes. (C) Higher magnification view of Ve in the lumen of the ER. (D) Tight apposition of inner vesicle (Ve) or tube (T) membrane and outer ER membrane gives the impression of “free” DMVs or tubes next to ER cisternae containing virus particles. (E) Projection of eight consecutive 1 nm tomogram slices of a high-pressure-frozen/freeze-substituted DENV-infected cell volume analyzed by ET. Stacked and single virus particles (Vi) are surrounded by a membrane that displays a spiky pattern resembling viral membrane proteins. Vi are inside membrane cisternae, which are tightly associated with the virus-induced vesicles (Ve).
Figure 3
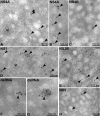
EM Localization of NS3 and E in DENV-Infected Cells (A–E) DENV-infected Huh-7 cells were fixed 24 hr postinfection, and thawed cryosections were labeled with antibodies specific for NS3 (A–C) or the glycoprotein E (D and E). Vesicles (Ve, arrowheads) into which cisternae of the ER (indicated) extend are shown in (A). The NS3-specific antibody significantly labels both Ve (arrowheads) and the CMs in (B). In (C), NS3-specific antibody labels a Ve (arrowhead), ER membranes, and the limiting membrane of a dilated ER cisterna containing virions (Vi). Dilated cisternae filled with Vi that are labeled with anti-E in (D), whereas the Ve (arrowheads) are not labeled. The E-specific antibody labels a single virion (Vi, arrowhead) within the lumen of an ER cisterna that is located within a site of Ve accumulation in (E). Note that the Ve appear as double-membraned spheres in thawed cryosections.
Figure 4
EM Localization of NS Proteins and DsRNA in DENV-Infected Cells (A–H) DENV-infected Huh-7 cells were fixed 24 hr postinfection, and thawed cryosections were labeled with antibodies against NS4A (A and B), NS4B (C), NS1 (D), NS2B (E), dsRNA, the presumed intermediate produced during DENV replication (F and G), or NS5 (H). All images show DENV-induced vesicles. All NS antibodies specifically label vesicles (arrowheads). Note that the virions (Vi) in (A) are not labeled with anti-NS4A and that the dsRNA-specific antibody (F and G) labels a discrete electron-dense structure located within (F) or outside (G) the vesicles.
Figure 5
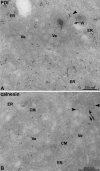
Virally Modified Membranes Are Derived from the ER (A and B) DENV-infected Huh-7 cells were fixed 24 hr postinfection, and thawed cryosections were labeled with PDI- (A) or calnexin-specific (B) antibodies. Arrowheads indicate anti-PDI or -calnexin labeling; arrows indicate virions (Vi) in the lumen of the ER. Note the ER cisternae in (A) that are abundantly labeled with PDI and that extend into the membrane structure containing vesicles (Ve) and CMs. Whereas PDI shows low but specific labeling of the vesicles in (A), calnexin specifically labels the CMs in (B).
Figure 6
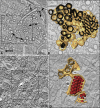
DENV-Induced Membrane Structures and ER Form a Continuous Intracellular Membrane Network (A) Huh-7 cells fixed at 26.5 hr postinfection were embedded in epoxy resin and analyzed by ET. Slice through a tomogram shows DENV-induced CMs, vesicles (Ve), and tubes (T) that form a network of interconnected membranes in continuity with the ER. Image represents an ∼9.5 nm thick slice (Z binning of eight 1.18 nm slices). (B) Three-dimensional surface model of the membranes in the boxed area in (A). The outer (cytosolic) face of the continuous membrane network is depicted in yellow; the ER lumen is dark. (C) Three nanometer slice through a tomogram (Z binning of three 0.998 nm slices). Stacked virus particles are seen in ER cisternae that are directly connected to virus-induced vesicles (white arrow). (D) Three-dimensional surface model of the virus-induced structures in the boxed area in (C) showing the continuity of virus-and vesicle-containing ER cisternae. ER membranes are depicted in yellow, inner vesicle membranes in light brown, and virus particles in red.
Figure 7
Characterization of DENV-Induced Vesicles and Viral Budding Sites and a Model of Their Possible Relation (A–C) Single slices of tomograms showing DENV-induced vesicles as invaginations of the ER membrane (white arrowheads). Note the diffuse electron density at the cytosolic face of the vesicle openings and the tight apposition of vesicle openings to the opposing ER/NE containing a virus particle (black arrowhead in [B]). The right panel in (C) shows a 3D surface rendering of the ER/vesicle continuity (yellow) and the tightly apposed ER (semitransparent) containing a virus particle (red). (D) Sixty nanometer section of the same sample as in (C), showing a vesicle in the ER and a putative virus budding event into the ER lumen located opposite the vesicle pore (arrow). (E) Single slice of a tomogram showing a vesicle in the ER, its pore (white arrowhead), and a putative budding site (arrow) opposite the vesicle pore. (F–I) Four slices through a tomogram, each ∼2 nm thick. Note the continuity of vesicle and ER membranes (white arrowheads) and the tight apposition of vesicle openings to the opposing NE. Virus particles (black arrowheads) are seen in the NE, in the ER, and in a cisterna close to the Golgi stack. (J) Three-dimensional surface rendering of ER and Golgi membranes (yellow), outer NE membrane (semitransparent), DENV-induced vesicles in the ER (light brown), virus particles (red), and a putative virus budding site (arrow) of the tomogram shown in (F–I). (K) The vesicle-containing ER segment shown in (J), rotated by 90° around the y axis, highlighting the vesicle openings. (L) Model of the relation between DENV replication, assembly, and virion release. Upon infection, the viral genome associates with the rough ER (ribosomes in blue), and the viral polyprotein, composed of the structural proteins (red) and the NS proteins (green), is synthesized on rough ER membranes. NS4A, together with other viral and perhaps cellular proteins, induces invaginations of the ER membrane, leading to the formation of vesicles that are connected to the cytosol via a pore. Inside these invaginations, RNA replication occurs. Viral capsid protein associates with progeny RNA genomes liberated through the pore-like structure into the cytosol. Virus budding occurs through the ER membrane located in close proximity to or opposite of the vesicles. Individual virions travel toward distal sites of the ER lumen, where they collect in dilated ER cisternae. They are transported, likely as individual virions, via secretory vesicles to the Golgi complex, where virion maturation occurs. Samples (A–I) were prepared as in Figure 6 and analyzed by ET, with the exception of (D), which was analyzed by TEM.
Similar articles
- Dengue virus- and hepatitis C virus-induced replication and assembly compartments: the enemy inside--caught in the web.
Chatel-Chaix L, Bartenschlager R. Chatel-Chaix L, et al. J Virol. 2014 Jun;88(11):5907-11. doi: 10.1128/JVI.03404-13. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623440 Free PMC article. Review. - Morphogenesis of Endoplasmic Reticulum Membrane-Invaginated Vesicles during Beet Black Scorch Virus Infection: Role of Auxiliary Replication Protein and New Implications of Three-Dimensional Architecture.
Cao X, Jin X, Zhang X, Li Y, Wang C, Wang X, Hong J, Wang X, Li D, Zhang Y. Cao X, et al. J Virol. 2015 Jun;89(12):6184-95. doi: 10.1128/JVI.00401-15. Epub 2015 Apr 1. J Virol. 2015. PMID: 25833056 Free PMC article. - Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells.
Junjhon J, Pennington JG, Edwards TJ, Perera R, Lanman J, Kuhn RJ. Junjhon J, et al. J Virol. 2014 May;88(9):4687-97. doi: 10.1128/JVI.00118-14. Epub 2014 Feb 12. J Virol. 2014. PMID: 24522909 Free PMC article. - Dengue Virus Selectively Annexes Endoplasmic Reticulum-Associated Translation Machinery as a Strategy for Co-opting Host Cell Protein Synthesis.
Reid DW, Campos RK, Child JR, Zheng T, Chan KWK, Bradrick SS, Vasudevan SG, Garcia-Blanco MA, Nicchitta CV. Reid DW, et al. J Virol. 2018 Mar 14;92(7):e01766-17. doi: 10.1128/JVI.01766-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29321322 Free PMC article. - Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories.
den Boon JA, Ahlquist P. den Boon JA, et al. Annu Rev Microbiol. 2010;64:241-56. doi: 10.1146/annurev.micro.112408.134012. Annu Rev Microbiol. 2010. PMID: 20825348 Review.
Cited by
- Subdomain cryo-EM structure of nodaviral replication protein A crown complex provides mechanistic insights into RNA genome replication.
Unchwaniwala N, Zhan H, Pennington J, Horswill M, den Boon JA, Ahlquist P. Unchwaniwala N, et al. Proc Natl Acad Sci U S A. 2020 Aug 4;117(31):18680-18691. doi: 10.1073/pnas.2006165117. Epub 2020 Jul 20. Proc Natl Acad Sci U S A. 2020. PMID: 32690711 Free PMC article. - A Combined Genetic-Proteomic Approach Identifies Residues within Dengue Virus NS4B Critical for Interaction with NS3 and Viral Replication.
Chatel-Chaix L, Fischl W, Scaturro P, Cortese M, Kallis S, Bartenschlager M, Fischer B, Bartenschlager R. Chatel-Chaix L, et al. J Virol. 2015 Jul;89(14):7170-86. doi: 10.1128/JVI.00867-15. Epub 2015 Apr 29. J Virol. 2015. PMID: 25926641 Free PMC article. - Role of host reticulon proteins in rearranging membranes for positive-strand RNA virus replication.
Diaz A, Ahlquist P. Diaz A, et al. Curr Opin Microbiol. 2012 Aug;15(4):519-24. doi: 10.1016/j.mib.2012.04.007. Epub 2012 May 21. Curr Opin Microbiol. 2012. PMID: 22621853 Free PMC article. Review. - Dengue Virus Reporter Replicon is a Valuable Tool for Antiviral Drug Discovery and Analysis of Virus Replication Mechanisms.
Kato F, Hishiki T. Kato F, et al. Viruses. 2016 May 5;8(5):122. doi: 10.3390/v8050122. Viruses. 2016. PMID: 27164125 Free PMC article. Review. - Development of antibody-based assays for high throughput discovery and mechanistic study of antiviral agents against yellow fever virus.
Gao Z, Zhang L, Ma J, Jurado A, Hong SH, Guo JT, Rice CM, MacDonald MR, Chang J. Gao Z, et al. Antiviral Res. 2020 Oct;182:104907. doi: 10.1016/j.antiviral.2020.104907. Epub 2020 Aug 14. Antiviral Res. 2020. PMID: 32798604 Free PMC article.
References
- Bartenschlager R., Miller S. Molecular aspects of Dengue virus replication. Future Microbiol. 2008;3:155–165. - PubMed
- Barth O.M. Replication of dengue viruses in mosquito cell cultures—a model from ultrastructural observations. Mem. Inst. Oswaldo Cruz. 1992;87:565–574. - PubMed
- Boulant S., Targett-Adams P., McLauchlan J. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 2007;88:2204–2213. - PubMed
- Buckley A., Gaidamovich S., Turchinskaya A., Gould E.A. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J. Gen. Virol. 1992;73:1125–1130. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources