Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion - PubMed (original) (raw)
Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion
Yadi Wu et al. Cancer Cell. 2009.
Abstract
The increased motility and invasiveness of tumor cells are reminiscent of epithelial-mesenchymal transition (EMT), which occurs during embryonic development, wound healing, and metastasis. In this study, we found that Snail is stabilized by the inflammatory cytokine TNFalpha through the activation of the NF-kappaB pathway. We demonstrated that NF-kappaB is required for the induction of COP9 signalosome 2 (CSN2), which, in turn, blocks the ubiquitination and degradation of Snail. Furthermore, we showed that the expression of Snail correlated with the activation of NF-kappaB in cancer cell lines and metastatic tumor samples. Knockdown of Snail expression inhibited cell migration and invasion induced by inflammatory cytokines and suppressed inflammation-mediated breast cancer metastasis. Our study provides a plausible mechanism for inflammation-induced metastasis.
Figures
Figure 1. The Inflammatory cytokine TNFα induced cell migration and invasion via protein stabilization of Snail
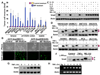
(A) The invasive ability of various cancer cell lines was examined by an invasion assay with regular culture medium (control medium) or macrophage conditioned medium (MP medium) as described in Materials and Methods. Error bars show standard deviation of three independent experiments in duplicate. (B) MCF7 and HEK293 GFP-Snail stable cell lines were cultured with control or MP medium overnight; the morphology and the intensity of GFP was examined under a fluorescent microscope. Scale bar = 100 µm. (C) Snail/HEK293 cells were treated with different cytokines (10 ng/ml for each) for 6 hr and Snail was analyzed by Western blotting. (D) HEK293 and breast cancer MCF7, T47D, and ZR75 Snail stable cells were treated with either TNFα or MG-132 (10 µM) for 6 hr and Snail was separated on 14% SDS-PAGE and examined by Western blotting. (E) Cells were treated with TNFα or MG-132 for 6 hr and endogenous Snail was analyzed by Western blotting as described in (D). (F) Snails were immunoprecipitated from Snail/HEK293 cells and the immunocomplex was then incubated with or without λ-phosphatase for 30 min and analyzed by western blotting. P and n indicate phosphorylated and unphosphorylated Snail, respectively. (G) Snail from Snail/HEK293 cells treated with TNFα for different time intervals was analyzed by Western blotting. (H) mRNA from MDA-MB231, treated with TNFα for different time intervals, was analyzed by RT-PCR. A reaction without template served as a negative control (Ng).
Figure 2. TNFα -mediated Snail stabilization was parallel with GSK-3β
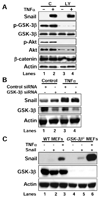
(A) Snail/HEK293 cells were pretreated with or without LY294002 (20 µM) for 1 hr followed by stimulation with or without TNFα for 6 hr. The expression of Snail and the activation of Akt, GSK-3β were examined by Western blotting. (B) Control and GSK-3β siRNA were expressed in Snail/HEK293 cells for 42 hr followed by treatment with or without TNFα for additional 6 hr. The expression of Snail, GSK-3β and actin was examined by Western blotting. (C) Snail was expressed in WT or _GSK-3β_−/− MEF cells for 42 hr followed by treatment with or without TNFα for additional 6 hr. The expression of Snail, GSK-3β and actin was examined by Western blotting.
Figure 3. Activation of NF-κB pathway was required for the stabilization of Snail
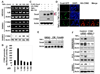
(A) Snail/HEK293 cells were pretreated with various inhibitors for 1 hr followed by stimulation with TNFα for 6 hr. The level of Snail was examined by Western blotting. (B) FLAG-tagged IKK or Myc-tagged p65 or mutant IκB was co-expressed with Snail in HEK293 cells that were treated with or without TNFα for 6 hr. Expression of Snail, IKK, p65, and IκB was analyzed by Western blotting. (C) GFP-Snail was co-expressed with p65 in HEK293 cells. After fixation, the cellular location of Snail (green) and p65 (red) was examined by immunofluorescent staining (nuclei were stained with DAPI; blue). Scale bar = 20 µm. (D) Control or p65 siRNA was expressed in Snail/HEK293, PC3, and HCT116 cells followed by TNFα or vehicle treatment for 6 hr. The levels of Snail and p65 were examined by Western blotting. (E) Snail was either expressed with or without p65 in _p65_−/− MEF cells treated with TNFα or vehicle for 6 hr. Snail and p65 was analyzed by Western blotting. (F) Snail and/or p65 were co-expressed with the E-cadherin promoter luciferase construct in HEK293 cells. After 40 hr, cells were treated with or without TNFα for 6 hr and luciferase activity was measured by using a Dual-Luciferase Reporter Assay (Promega) (mean ± SD in three separate experiments).
Figure 4. Transcriptional dependence of NF-κB was involved in the stabilization of Snail
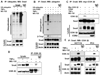
(A) Wild-type (WT) or various deletion mutants of p65 (scheme diagram shown on left panel) were co-expressed with Snail in HEK293 cells and the expression of Snail, IκB, and p65 was analyzed by Western blotting. (B) Snail/HEK293 cells were pretreated with or without actinomycin D (Act D; 0.5 µg/ml) for 1 hr followed by TNFα stimulation for 6 hr. Snail or IκB was analyzed by Western blotting. (C) After stimulation with TNFα for 6 hr, Snail/HEK293 cells were washed twice with fresh medium to remove residual TNFα followed by incubation without or with Act D in combination with TNFα or TNFα /MG-132 together over a time course. Snail or IκB was analyzed by Western blotting. (D) Densitometry results for the level of Snail described above were plotted.
Figure 5. CSN2 induced Snail stabilization
(A) HEK293 and SKBR3 cells were treated with TNFα for different time intervals and mRNA from these cells was analyzed by semi-quantitative RT-PCR. Expression of CSN2 was also analyzed in SKBR3 cells. (B) The CSN2 promoter luciferase construct was co-expressed with WT or different deletion mutants of p65 in HEK293 cells. Luciferase activities were normalized and determined (mean ± SD in three separate experiments). (C) Snail/HEK293 cells were either treated with TNFα or MG-132 for 6 hr or co-expressed with HA-CSN2. Expression of Snail and CSN2 was separated on 14% SDS-PAGE and analyzed by Western blotting. (D) GFP-Snail was co-expressed with CSN2 or empty vector in HEK293 cells followed by treatment with TNFα or vehicle for 6 hr. The cellular location of Snail (green) and CSN2 (red) was examined by immunofluorescent staining (nuclei were stained with DAPI; blue). Scale bar = 100 µm. (E) Snail/HEK293 cells were pretreated with either DMSO, or Curcumin (CM; 50 nM) or Emodin (400 nM) for 1 hr followed by TNFα for 6 hr. Snail was examined by Western blotting. (F) CSN2 siRNA was expressed in Snail/HEK293, PC3, and SKBR3 cells followed by treatment with TNFα or vehicle. Snail and CSN2 were examined by Western blotting.
Figure 6. CSN2 inhibited the ubiquitination of Snail
(A) HA-ubiquitin was expressed in Snail/HEK293 cells followed by treatment with either TNFα or MG-132 for 6 hr. Ubiquitin was immunoprecipitated from equal amount of lysates (two lower panels) and the ubiquitinated Snail was examined by Western blotting. (B) HA-ubiquitin was co-expressed with HA-CSN2 or control vector in Snail/HEK293 cells followed by treatment with TNFα or MG-132 for 6 hr. After Snail was immunoprecipitated, the ubiquitination of Snail and the bound β-Trcp were examined by Western blotting. (C) Myc-GSK-3β was co-expressed with HA-CSN2 or control vector in Snail/HEK293 cells for 42 hr followed by treatment with MG-132 or TNFα for additional 6 h. Snail was immunoprecipitated and the association of GSK-3β was examined by Western blotting. (D) HCT116 cells were either treated with TNFα or MG-132. Endogenous GSK-3β was immunoprecipitated and the bound endogenous Snail was detected by Western blotting. (E) Cells were treated with TNFα or MG-132. Endogenous Snail was immunoprecipitated and the associated endogenous GSK-3β was detected by Western blotting.
Figure 7. Knockdown of Snail expression suppressed inflammation-induced cell migration and invasion in vitro
(A) In the top two panels, mRNA from MDA-MB231 and MDA-MB435 cells and their corresponding stable transfectants with knockdown of Snail expression were analyzed by RT-PCR. In the bottom five panels, expression of Snail (treated with MG-132 for 6 hr), N-cadherin, ZO-1, Claudin-1, and actin was analyzed by Western blotting. (B) The motile behavior of MDA-MB231 and its corresponding Snail knockdown cells was analyzed by wound healing assay. The percentage of migrating cells was shown (mean ± SEM in three separate experiments) and the representative images were shown in Figure S12. (C) The invasive ability of MDA-MB231 and MDA-MB435 cells and their corresponding stable transfectants with knockdown of Snail expression was analyzed by the invasion assay. The representative images were shown and the percentage of invasive cells was plotted in bottom graph (mean ± SEM in three separate experiments). Scale bar = 200 µm.
Figure 8. Knockdown of Snail expression suppressed inflammation-induced metastasis in vivo
(A) MDA-MB435 and MDA-MB231 cells and their corresponding stable transfectants with knockdown of Snail expression were injected into the tail vein of ICR-SCID mice. After 6–9 wks, the number of lung metastases was plotted (mean ± SEM, n = 6) and the representative photography of lung was shown. For lung metastases, *P < 0.01 for parental cells compared with their stable transfectants with knockdown of Snail expression after treatment with LPS, #P < 0.05 when compared with PBS stimulation for parental cells, †P < 0.05 for parental cells compared with their stable transfectants with knockdown of Snail expression treated with PBS. (B) Nuclear extracts were prepared from various cancer cell lines and the expression of Snail and p65 was analyzed by Western blotting. Poly (ADP-ribose) polymerase (PARP) was used as a control for the equal loading of nuclear extracts. (C) The 110 surgical specimens of breast cancer were immuno-stained using antibodies to Snail, p65, as well as the control serum (data not shown). Representative stainings from the same tumor samples are shown. Scale bar = 100 µm. (D) A proposed model to illustrate the stabilization of Snail by TNFα leading to EMT and metastasis.
Comment in
- Inflamed snail speeds metastasis.
Yang CC, Wolf DA. Yang CC, et al. Cancer Cell. 2009 May 5;15(5):355-7. doi: 10.1016/j.ccr.2009.04.003. Cancer Cell. 2009. PMID: 19411063
Similar articles
- Inflamed snail speeds metastasis.
Yang CC, Wolf DA. Yang CC, et al. Cancer Cell. 2009 May 5;15(5):355-7. doi: 10.1016/j.ccr.2009.04.003. Cancer Cell. 2009. PMID: 19411063 - Gemifloxacin inhibits migration and invasion and induces mesenchymal-epithelial transition in human breast adenocarcinoma cells.
Chen TC, Hsu YL, Tsai YC, Chang YW, Kuo PL, Chen YH. Chen TC, et al. J Mol Med (Berl). 2014 Jan;92(1):53-64. doi: 10.1007/s00109-013-1083-4. Epub 2013 Sep 5. J Mol Med (Berl). 2014. PMID: 24005829 - Stabilization of Snail through AKT/GSK-3β signaling pathway is required for TNF-α-induced epithelial-mesenchymal transition in prostate cancer PC3 cells.
Wang H, Fang R, Wang XF, Zhang F, Chen DY, Zhou B, Wang HS, Cai SH, Du J. Wang H, et al. Eur J Pharmacol. 2013 Aug 15;714(1-3):48-55. doi: 10.1016/j.ejphar.2013.05.046. Epub 2013 Jun 11. Eur J Pharmacol. 2013. PMID: 23769744 - The activated NF-kappaB-Snail-RKIP circuitry in cancer regulates both the metastatic cascade and resistance to apoptosis by cytotoxic drugs.
Wu K, Bonavida B. Wu K, et al. Crit Rev Immunol. 2009;29(3):241-54. doi: 10.1615/critrevimmunol.v29.i3.40. Crit Rev Immunol. 2009. PMID: 19538137 Review. - Dual role of NO donors in the reversal of tumor cell resistance and EMT: Downregulation of the NF-κB/Snail/YY1/RKIP circuitry.
Bonavida B, Baritaki S. Bonavida B, et al. Nitric Oxide. 2011 Jan 1;24(1):1-7. doi: 10.1016/j.niox.2010.10.001. Epub 2010 Oct 8. Nitric Oxide. 2011. PMID: 20933602 Review.
Cited by
- Interleukin 6 and cancer resistance in glioblastoma multiforme.
Detchou D, Barrie U. Detchou D, et al. Neurosurg Rev. 2024 Sep 5;47(1):541. doi: 10.1007/s10143-024-02783-5. Neurosurg Rev. 2024. PMID: 39231832 Review. - The role of phenotypic plasticity in the escape of cancer cells from targeted therapy.
Emmons MF, Faião-Flores F, Smalley KSM. Emmons MF, et al. Biochem Pharmacol. 2016 Dec 15;122:1-9. doi: 10.1016/j.bcp.2016.06.014. Epub 2016 Jun 25. Biochem Pharmacol. 2016. PMID: 27349985 Free PMC article. Review. - Systematic analysis reveals a functional role for STAMBPL1 in the epithelial-mesenchymal transition process across multiple carcinomas.
Ambroise G, Yu TT, Zhang B, Kacal M, Hao Y, Queiroz AL, Ouchida AT, Lindskog C, Norberg E, Vakifahmetoglu-Norberg H. Ambroise G, et al. Br J Cancer. 2020 Sep;123(7):1164-1177. doi: 10.1038/s41416-020-0972-x. Epub 2020 Jul 8. Br J Cancer. 2020. PMID: 32636467 Free PMC article. - EMT or EMT-Promoting Transcription Factors, Where to Focus the Light?
Ansieau S, Collin G, Hill L. Ansieau S, et al. Front Oncol. 2014 Dec 16;4:353. doi: 10.3389/fonc.2014.00353. eCollection 2014. Front Oncol. 2014. PMID: 25566496 Free PMC article. Review. No abstract available. - Twist-mediated PAR1 induction is required for breast cancer progression and metastasis by inhibiting Hippo pathway.
Wang Y, Liao R, Chen X, Ying X, Chen G, Li M, Dong C. Wang Y, et al. Cell Death Dis. 2020 Jul 9;11(7):520. doi: 10.1038/s41419-020-2725-4. Cell Death Dis. 2020. PMID: 32647142 Free PMC article.
References
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. - PubMed
- Berse M, Bounpheng M, Huang X, Christy B, Pollmann C, Dubiel W. Ubiquitin-dependent degradation of Id1 and Id3 is mediated by the COP9 signalosome. J Mol Biol. 2004;343:361–370. - PubMed
- Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J, Nieto MA. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 2002;21:3241–3246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-CA125454/CA/NCI NIH HHS/United States
- T32CA117834/CA/NCI NIH HHS/United States
- R01 CA104748/CA/NCI NIH HHS/United States
- R01 DK048498/DK/NIDDK NIH HHS/United States
- R01 DK48498/DK/NIDDK NIH HHS/United States
- R01 CA125454/CA/NCI NIH HHS/United States
- T32 CA117834/CA/NCI NIH HHS/United States
- R01 CA125454-02/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials