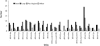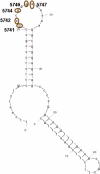The diversity present in 5140 human mitochondrial genomes - PubMed (original) (raw)
The diversity present in 5140 human mitochondrial genomes
Luísa Pereira et al. Am J Hum Genet. 2009 May.
Abstract
We analyzed the current status (as of the end of August 2008) of human mitochondrial genomes deposited in GenBank, amounting to 5140 complete or coding-region sequences, in order to present an overall picture of the diversity present in the mitochondrial DNA of the global human population. To perform this task, we developed mtDNA-GeneSyn, a computer tool that identifies and exhaustedly classifies the diversity present in large genetic data sets. The diversity observed in the 5140 human mitochondrial genomes was compared with all possible transitions and transversions from the standard human mitochondrial reference genome. This comparison showed that tRNA and rRNA secondary structures have a large effect in limiting the diversity of the human mitochondrial sequences, whereas for the protein-coding genes there is a bias toward less variation at the second codon positions. The analysis of the observed amino acid variations showed a tolerance of variations that convert between the amino acids V, I, A, M, and T. This defines a group of amino acids with similar chemical properties that can interconvert by a single transition.
Figures
Figure 1
The Geographical Origin of the 5140 mtDNA Sequences Deposited in GenBank, as of the End of August 2008 African-American descendants were included in sub-Saharan Africa and European-American descendants were included in Eurasians given that they were homogeneous groups, bearing haplogroups belonging to these regions, whereas USA “Hispanics” constituted a mixed sample, with most lineages belonging to East Asian haplogroups observed in Native Americans.
Figure 2
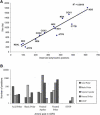
Protein Diversity Correlation between the observed polymorphic positions and the gene size (bp) in the 13 protein-coding genes (A) and distributions of types of amino acids before and after the substitution (B).
Figure 3
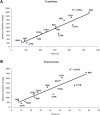
Transitions and Transversions Correlation between the observed and maximum possible variations for (A) transitions and (B) transversions in the 13 protein-coding genes.
Figure 4
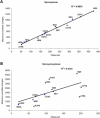
Synonymous and Nonsynonymous Sequence Changes Correlation between observed and maximum possible variations for (A) synonymous and (B) nonsynonymous substitutions in the 13 protein-coding genes.
Figure 5
Correlation between Observed and Maximum Possible Variations for Amino Acids Originated from Nonsynonymous Mutations The amino acids from group (VIAMT) are represented by red squares and the ones from group (FLPS) are represented by blue circles; the remaining amino acids are represented by black diamonds.
Figure 6
Distribution of Polymorphic Positions in the Various Structural Regions of the tRNA Genes
Figure 7
Variability in the Region of the Origin of the Light Strand Related to the Inferred DNA Secondary Structure Secondary structure of the OL region inferred in mfold with the observed polymorphic positions 5741, 5742, 5744, 5746, and 5747 indicated.
Figure 8
Variability in a tRNA Gene Related to the RNA Secondary Structure The secondary structure of the threonine tRNA and the observed polymorphic positions in the human database (gray hexagons), and positions that are 100% (square) and >90% (circle) conserved in mammalian species.
Comment in
- mtDNA data mining in GenBank needs surveying.
Yao YG, Salas A, Logan I, Bandelt HJ. Yao YG, et al. Am J Hum Genet. 2009 Dec;85(6):929-33; author reply 933. doi: 10.1016/j.ajhg.2009.10.023. Am J Hum Genet. 2009. PMID: 20004768 Free PMC article. No abstract available.
Similar articles
- Complete mitochondrial DNA sequence of oyster Crassostrea hongkongensis-a case of "Tandem duplication-random loss" for genome rearrangement in Crassostrea?
Yu Z, Wei Z, Kong X, Shi W. Yu Z, et al. BMC Genomics. 2008 Oct 11;9:477. doi: 10.1186/1471-2164-9-477. BMC Genomics. 2008. PMID: 18847502 Free PMC article. - The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization.
Crozier RH, Crozier YC. Crozier RH, et al. Genetics. 1993 Jan;133(1):97-117. doi: 10.1093/genetics/133.1.97. Genetics. 1993. PMID: 8417993 Free PMC article. - Mitochondrial DNA of Clathrina clathrus (Calcarea, Calcinea): six linear chromosomes, fragmented rRNAs, tRNA editing, and a novel genetic code.
Lavrov DV, Pett W, Voigt O, Wörheide G, Forget L, Lang BF, Kayal E. Lavrov DV, et al. Mol Biol Evol. 2013 Apr;30(4):865-80. doi: 10.1093/molbev/mss274. Epub 2012 Dec 6. Mol Biol Evol. 2013. PMID: 23223758 - Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA.
Burger G, Zhu Y, Littlejohn TG, Greenwood SJ, Schnare MN, Lang BF, Gray MW. Burger G, et al. J Mol Biol. 2000 Mar 24;297(2):365-80. doi: 10.1006/jmbi.2000.3529. J Mol Biol. 2000. PMID: 10715207
Cited by
- Divorcing the Late Upper Palaeolithic demographic histories of mtDNA haplogroups M1 and U6 in Africa.
Pennarun E, Kivisild T, Metspalu E, Metspalu M, Reisberg T, Moisan JP, Behar DM, Jones SC, Villems R. Pennarun E, et al. BMC Evol Biol. 2012 Dec 3;12:234. doi: 10.1186/1471-2148-12-234. BMC Evol Biol. 2012. PMID: 23206491 Free PMC article. - 60,000 years of interactions between Central and Eastern Africa documented by major African mitochondrial haplogroup L2.
Silva M, Alshamali F, Silva P, Carrilho C, Mandlate F, Jesus Trovoada M, Černý V, Pereira L, Soares P. Silva M, et al. Sci Rep. 2015 Jul 27;5:12526. doi: 10.1038/srep12526. Sci Rep. 2015. PMID: 26211407 Free PMC article. - MitoLSDB: a comprehensive resource to study genotype to phenotype correlations in human mitochondrial DNA variations.
K S, Jalali S, Scaria V, Bhardwaj A. K S, et al. PLoS One. 2013 Apr 9;8(4):e60066. doi: 10.1371/journal.pone.0060066. Print 2013. PLoS One. 2013. PMID: 23585830 Free PMC article. - From Caves to the Savannah, the Mitogenome History of Modern Lions (Panthera leo) and Their Ancestors.
Broggini C, Cavallini M, Vanetti I, Abell J, Binelli G, Lombardo G. Broggini C, et al. Int J Mol Sci. 2024 May 10;25(10):5193. doi: 10.3390/ijms25105193. Int J Mol Sci. 2024. PMID: 38791233 Free PMC article.
References
- Ingman M., Kaessmann H., Pääbo S., Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. - PubMed
- De Benedictis G., Rose G., Carreiri G., De Luca M., Falcone E., Passarino G., Bonafe M., Monti D., Baggio G., Bertolini S. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources