Non-random reassortment in human influenza A viruses - PubMed (original) (raw)
Non-random reassortment in human influenza A viruses
Raul Rabadan et al. Influenza Other Respir Viruses. 2008 Jan.
Abstract
Background: The influenza A virus has two basic modes of evolution. Because of a high error rate in the process of replication by RNA polymerase, the viral genome drifts via accumulated mutations. The second mode of evolution is termed a shift, which results from the reassortment of the eight segments of this virus. When two different influenza viruses co-infect the same host cell, new virions can be released that contain segments from both parental strains. This type of shift has been the source of at least two of the influenza pandemics in the 20th century (H2N2 in 1957 and H3N2 in 1968).
Objectives: The methods to measure these genetic shifts have not yet provided a quantitative answer to questions such as: what is the rate of genetic reassortment during a local epidemic? Are all possible reassortments equally likely or are there preferred patterns?
Methods: To answer these questions and provide a quantitative way to measure genetic shifts, a new method for detecting reassortments from nucleotide sequence data was created that does not rely upon phylogenetic analysis. Two different sequence databases were used: human H3N2 viruses isolated in New York State between 1995 and 2006, and human H3N2 viruses isolated in New Zealand between 2000 and 2005.
Results: Using this new method, we were able to reproduce all the reassortments found in earlier works, as well as detect, with very high confidence, many reassortments that were not detected by previous authors. We obtain a lower bound on the reassortment rate of 2-3 events per year, and find a clear preference for reassortments involving only one segment, most often hemagglutinin or neuraminidase. At a lower frequency several segments appear to reassort in vivo in defined groups as has been suggested previously in vitro.
Conclusions: Our results strongly suggest that the patterns of reassortment in the viral population are not random. Deciphering these patterns can be a useful tool in attempting to understand and predict possible influenza pandemics.
Figures
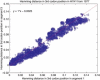
Figure 1
Hamming distance in third codon position between human H1N1 influenza strains (237 cases of human H1N1 from 1977 to 2006) in segment 1 versus segment 3. The red line is the best line fit to the data.
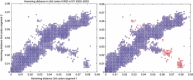
Figure 2
Hamming distance in third codon position between human H3N2 influenza strains in the New York state (208 sequences from 2000 to 2003) in segment 1 versus segment 2. In red are the pairs of sequences that contain A/New York/11/2003(H3N2).
Figure 3
Hamming distance in third codon position between human H3N2 influenza strains in the New York state (208 sequences from 2000 to 2003) in segment 1 versus segment 2. If we create new segments by randomly permuting the third codon positions of both segments together, we create a new distribution (in red is one example). If there are no reassortments the permuted distribution and the real one should be similar (around the diagonal).
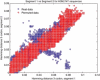
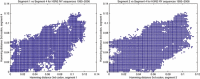
Figure 4
The evolutionary rate in different segments is similar if we take into account the changes in third codon positions. In this figure is represented the Hamming distance in third codon position between every pair of viruses found in New York State between 1995 and 2006 (581 sequences). On the left is shown the distance in third codon position in segment 1 versus segment 4, and on the right segment 2 versus segment 4. It can be appreciated that most of the points are along the slope 1 line, indicating similar rates between the different segments.
Similar articles
- Seasonal H3N2 and 2009 Pandemic H1N1 Influenza A Viruses Reassort Efficiently but Produce Attenuated Progeny.
Phipps KL, Marshall N, Tao H, Danzy S, Onuoha N, Steel J, Lowen AC. Phipps KL, et al. J Virol. 2017 Aug 10;91(17):e00830-17. doi: 10.1128/JVI.00830-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28637755 Free PMC article. - Parallel evolution between genomic segments of seasonal human influenza viruses reveals RNA-RNA relationships.
Jones JE, Le Sage V, Padovani GH, Calderon M, Wright ES, Lakdawala SS. Jones JE, et al. Elife. 2021 Aug 27;10:e66525. doi: 10.7554/eLife.66525. Elife. 2021. PMID: 34448455 Free PMC article. - A phylogenetic approach to detecting reassortments in viruses with segmented genomes.
Suzuki Y. Suzuki Y. Gene. 2010 Sep 15;464(1-2):11-6. doi: 10.1016/j.gene.2010.05.002. Epub 2010 May 28. Gene. 2010. PMID: 20546849 - [Molecular characterization of human influenza viruses--a look back on the last 10 years].
Schweiger B. Schweiger B. Berl Munch Tierarztl Wochenschr. 2006 Mar-Apr;119(3-4):167-78. Berl Munch Tierarztl Wochenschr. 2006. PMID: 16573207 Review. German. - [The pandemic influenza virus H1N1/2009: a review of the molecular biology, phylogeny, history of reassortments, and parameters of host switching].
Stech J, Beer M, Vahlenkamp T, Harder T. Stech J, et al. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2010 Dec;53(12):1231-7. doi: 10.1007/s00103-010-1166-0. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2010. PMID: 21161472 Review. German.
Cited by
- Reassortment Network of Influenza A Virus.
Gong X, Hu M, Chen W, Yang H, Wang B, Yue J, Jin Y, Liang L, Ren H. Gong X, et al. Front Microbiol. 2021 Dec 16;12:793500. doi: 10.3389/fmicb.2021.793500. eCollection 2021. Front Microbiol. 2021. PMID: 34975817 Free PMC article. - HopPER: an adaptive model for probability estimation of influenza reassortment through host prediction.
Yin R, Zhou X, Rashid S, Kwoh CK. Yin R, et al. BMC Med Genomics. 2020 Jan 23;13(1):9. doi: 10.1186/s12920-019-0656-7. BMC Med Genomics. 2020. PMID: 31973709 Free PMC article. - Intrasubtype reassortments cause adaptive amino acid replacements in H3N2 influenza genes.
Neverov AD, Lezhnina KV, Kondrashov AS, Bazykin GA. Neverov AD, et al. PLoS Genet. 2014 Jan;10(1):e1004037. doi: 10.1371/journal.pgen.1004037. Epub 2014 Jan 9. PLoS Genet. 2014. PMID: 24415946 Free PMC article. - Reassortment between two serologically unrelated bluetongue virus strains is flexible and can involve any genome segment.
Shaw AE, Ratinier M, Nunes SF, Nomikou K, Caporale M, Golder M, Allan K, Hamers C, Hudelet P, Zientara S, Breard E, Mertens P, Palmarini M. Shaw AE, et al. J Virol. 2013 Jan;87(1):543-57. doi: 10.1128/JVI.02266-12. Epub 2012 Oct 24. J Virol. 2013. PMID: 23097432 Free PMC article. - RNA Virus Reassortment: An Evolutionary Mechanism for Host Jumps and Immune Evasion.
Vijaykrishna D, Mukerji R, Smith GJ. Vijaykrishna D, et al. PLoS Pathog. 2015 Jul 9;11(7):e1004902. doi: 10.1371/journal.ppat.1004902. eCollection 2015 Jul. PLoS Pathog. 2015. PMID: 26158697 Free PMC article. Review. No abstract available.
References
- Schweiger B, Bruns L, Meixenberger K. Reassortment between human A(H3N2) viruses is an important evolutionary mechanism. Vaccine 2006; 24:6683–6690, Epub June 16, 2006. - PubMed
- Barr IG, Komadina N, Hurt A et al. Reassortants in recent human influenza A and B isolates from South East Asia and Oceania. Virus Res 2003; 98:35–44. - PubMed
- Lindstrom SE, Cox NJ, Klimov A. Genetic analysis of human H2N2 and early H3N2 influenza viruses, 1957–1972: evidence for genetic divergence and multiple reassortment events. Virology 2004; 328:101–119. - PubMed
- Lubeck MD, Palese P, Schulman JL. Nonrandom association of parental genes in influenza A virus recombinants. Virology 1979; 95:269–274. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical