miR-145 and miR-143 regulate smooth muscle cell fate and plasticity - PubMed (original) (raw)
miR-145 and miR-143 regulate smooth muscle cell fate and plasticity
Kimberly R Cordes et al. Nature. 2009.
Abstract
MicroRNAs (miRNAs) are regulators of myriad cellular events, but evidence for a single miRNA that can efficiently differentiate multipotent stem cells into a specific lineage or regulate direct reprogramming of cells into an alternative cell fate has been elusive. Here we show that miR-145 and miR-143 are co-transcribed in multipotent murine cardiac progenitors before becoming localized to smooth muscle cells, including neural crest stem-cell-derived vascular smooth muscle cells. miR-145 and miR-143 were direct transcriptional targets of serum response factor, myocardin and Nkx2-5 (NK2 transcription factor related, locus 5) and were downregulated in injured or atherosclerotic vessels containing proliferating, less differentiated smooth muscle cells. miR-145 was necessary for myocardin-induced reprogramming of adult fibroblasts into smooth muscle cells and sufficient to induce differentiation of multipotent neural crest stem cells into vascular smooth muscle. Furthermore, miR-145 and miR-143 cooperatively targeted a network of transcription factors, including Klf4 (Kruppel-like factor 4), myocardin and Elk-1 (ELK1, member of ETS oncogene family), to promote differentiation and repress proliferation of smooth muscle cells. These findings demonstrate that miR-145 can direct the smooth muscle fate and that miR-145 and miR-143 function to regulate the quiescent versus proliferative phenotype of smooth muscle cells.
Figures
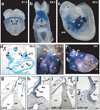
Figure 1. miR-143 and miR-145 are cardiac and smooth muscle-specific miRNAs
(a–c) Whole mounts showing cardiac-specific β-gal activity in transgenic mouse embryos with 4.2 kb enhancer-lacZ construct (Supplementary Fig. 2a) at indicated time points. (d) Transverse section of (c) showing β-gal expression in pharyngeal mesoderm (pm), pharyngeal endoderm (pe), dorsal aorta (da), myocardium (mc), endocardium (ec). (e,f) β-gal expression in embryonic day (E) 15.5 (e) or post-natal (P21) (f) heart. Ao, aorta; pa, pulmonary artery. (g,h) Transverse sections of (f); co, coronary artery. (i,j) Section in-situ hybridization of miR-145 in P21 heart section. (h) and (j) represent higher magnification of boxed areas. (pcm, precardiac mesoderm; ht, heart; h, head; ot, outflow tract; rv, right ventricle; lv, left ventricle; cv, cardinal vein; ra, right atrium; la, left atrium).
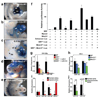
Figure 2. SRF and Nkx2.5 directly regulate cardiac and smooth muscle expression of miR-143 and miR-145
(a–e) Lateral (a, b, c, e) or frontal (d) cardiac views of transgenic embryos containing indicated lacZ constructs and stained for β-Gal activity. (f) Fold-activation of luciferase activity directed by introduction of SRF, Myocd or Nkx2.5 expression vectors with the miR-143/145 enhancer in Cos cells. All changes were statistically significant (n=5). (g) miR-143 and miR-145 expression levels assessed by qPCR day 10 embyroid bodies (EBs) of indicated genotypes. (h) qPCR of miR-143 and miR-145 in Nkx2.5+/− and _Nkx2.5_−/− E9.5 hearts relative to WT. (i,j) qPCR of miRNAs in injured vessels (i) or atherosclerotic lesions (j) compared to normal arterial expression. Results shown in (f_–_j) are the average of three experiments. (ot, outflow tract; ra, right atrium; lv, left ventricle; rv, right ventricle; la, left atrium; dorsal aorta). *, p<0.05. Error bars indicate SD.
Figure 3. miR-145 directs vascular smooth muscle cell fate
(a) Immunocytochemistry of 10T1/2 fibroblasts using smooth muscle (Sm) α-actin antibodies (red) under conditions indicated; nuclear stain, Dapi (blue). (b) Quantification of Sm-α-actin positive cells (n=6). (c) qPCR of Sm gene expression in fibroblasts transfected with Myocd with or without anti-miR-145 or (d) fibroblasts transfected with 50 ng Myocd with or without miR-145 (n=5). (e) Western blot of calponin and Sm-α-actin. (f) Immunocytochemistry of neural crest stem cells (Joma1.3 NCCs) with or without miR-145 using antibodies indicated (green); tamoxifen (4OHT) was removed to allow differentiation. Quantification of percent Sm-α-actin + cells relative to total Dapi+ nuclei (blue) (n=6). (g) qPCR of Sm gene expression in NCCs with miR-145 expression (n=5); p75 is a marker of undifferentiated neural crest cells. (h) Western blot of Sm-α-actin and calponin. (i) Calcium flux [Ca2+]i in SMCs derived from NCCs or rat aortic SMCs in response to endothelin-1 (Et-1) stimulation at 30 sec. Error bars indicating SD. *, p<0.05.
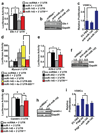
Figure 4. miR-143 and miR-145 target a network of factors to promote VSMC differentiation and repress proliferation
(a) Luciferase activity in Cos cells upon introduction of Elk-1 3′ UTR or mutant 3′ UTR (mut) downstream of a CMV-driven luciferase reporter with indicated miRNAs (n=5). (b) Elk-1 protein in cell lysates from A10 VSMCs transfected with a scrambled (scr) miRNA or anti-miR-143 or -miR-145 assessed by western blot. (c) Proliferation of VSMCs upon inhibition of miR-143 or miR-145 relative to control (5%FBS) (n=5). (d) Luciferase activity in Cos cells with Myocd 3′ UTR sequences with indicated miRNAs (n=5). The Myocd binding site (BS) was mutated in the context of a 4x concatemer. (e) Luciferase activity with wt or mutated _Klf4_-3′ UTR upon introduction of indicated miRNAs (n=5). (f) Analysis of Klf4 protein in cell lysates from A10 cells transfected with indicated anti-miRs by western blot. (g) Luciferase activity of wt or mutated _CamkII_-δ3′ UTR (n=5). (h) Western analysis for CamkII-δ protein in A10 cells transfected with scr miRNA or anti-miR-145. (i) Proliferation of VSMCs relative to control (n=5). Error bars indicate SD. Densitometry calculation performed by Image J. *, p<0.05.
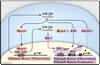
Figure 5. Model of miR-143 and miR-145 regulation of smooth muscle cell proliferation and differentiation
miR-143 and miR-145 are positively regulated by SRF and function to repress multiple factors that normally promote the less differentiated, more proliferative smooth muscle phenotype (purple). These include Klf4, which also represses Myocd. miR-145 has a positive effect on Myocd activity to concurrently promote the more differentiated smooth muscle phenotype (pink), thereby also functioning to reinforce its own expression. Effects of miR-145 and miR-143 converge on SRF-dependent transcription by regulation of co-activators and co-repressors to dictate the proliferative or differentiated phenotype of VSMCs. Positive regulation of Myocd by miR-145 results in reinforcement of miR-145 and miR-143 expression and the differentiated phenotype. Dashed lines indicate indirect effects.
Similar articles
- The role of microRNA-145 in human embryonic stem cell differentiation into vascular cells.
Yamaguchi S, Yamahara K, Homma K, Suzuki S, Fujii S, Morizane R, Monkawa T, Matsuzaki Y, Kangawa K, Itoh H. Yamaguchi S, et al. Atherosclerosis. 2011 Dec;219(2):468-74. doi: 10.1016/j.atherosclerosis.2011.09.004. Epub 2011 Sep 9. Atherosclerosis. 2011. PMID: 21945499 - MicroRNA MiR-199a-5p regulates smooth muscle cell proliferation and morphology by targeting WNT2 signaling pathway.
Hashemi Gheinani A, Burkhard FC, Rehrauer H, Aquino Fournier C, Monastyrskaya K. Hashemi Gheinani A, et al. J Biol Chem. 2015 Mar 13;290(11):7067-86. doi: 10.1074/jbc.M114.618694. Epub 2015 Jan 16. J Biol Chem. 2015. PMID: 25596533 Free PMC article. - Serotonin augments smooth muscle differentiation of bone marrow stromal cells.
Hirota N, McCuaig S, O'Sullivan MJ, Martin JG. Hirota N, et al. Stem Cell Res. 2014 May;12(3):599-609. doi: 10.1016/j.scr.2014.02.003. Epub 2014 Feb 13. Stem Cell Res. 2014. PMID: 24595007 - The mechanism of stem cell differentiation into smooth muscle cells.
Xiao Q, Wang G, Luo Z, Xu Q. Xiao Q, et al. Thromb Haemost. 2010 Sep;104(3):440-8. doi: 10.1160/TH09-11-0794. Epub 2010 Jun 10. Thromb Haemost. 2010. PMID: 20539914 Review. - Epigenetics and miRNA emerge as key regulators of smooth muscle cell phenotype and function.
Clifford RL, Singer CA, John AE. Clifford RL, et al. Pulm Pharmacol Ther. 2013 Feb;26(1):75-85. doi: 10.1016/j.pupt.2012.07.002. Epub 2012 Jul 16. Pulm Pharmacol Ther. 2013. PMID: 22800879 Free PMC article. Review.
Cited by
- BMP2 promotes the differentiation of neural stem cells into dopaminergic neurons in vitro via miR-145-mediated upregulation of Nurr1 expression.
Yan W, Chen ZY, Chen JQ, Chen HM. Yan W, et al. Am J Transl Res. 2016 Sep 15;8(9):3689-3699. eCollection 2016. Am J Transl Res. 2016. PMID: 27725851 Free PMC article. - miRNA-dysregulation associated with tenderness variation induced by acute stress in Angus cattle.
Zhao C, Tian F, Yu Y, Liu G, Zan L, Updike MS, Song J. Zhao C, et al. J Anim Sci Biotechnol. 2012 Jun 1;3(1):12. doi: 10.1186/2049-1891-3-12. J Anim Sci Biotechnol. 2012. PMID: 22958451 Free PMC article. - Micromanaging abdominal aortic aneurysms.
Maegdefessel L, Spin JM, Adam M, Raaz U, Toh R, Nakagami F, Tsao PS. Maegdefessel L, et al. Int J Mol Sci. 2013 Jul 11;14(7):14374-94. doi: 10.3390/ijms140714374. Int J Mol Sci. 2013. PMID: 23852016 Free PMC article. Review. - MEF2C-MYOCD and Leiomodin1 Suppression by miRNA-214 Promotes Smooth Muscle Cell Phenotype Switching in Pulmonary Arterial Hypertension.
Sahoo S, Meijles DN, Al Ghouleh I, Tandon M, Cifuentes-Pagano E, Sembrat J, Rojas M, Goncharova E, Pagano PJ. Sahoo S, et al. PLoS One. 2016 May 4;11(5):e0153780. doi: 10.1371/journal.pone.0153780. eCollection 2016. PLoS One. 2016. PMID: 27144530 Free PMC article. - The role of miR-143/miR-145 in the development, diagnosis, and treatment of diabetes.
Rasmi Y, Mohamed YA, Alipour S, Ahmed S, Abdelmajed SS. Rasmi Y, et al. J Diabetes Metab Disord. 2023 Oct 11;23(1):39-47. doi: 10.1007/s40200-023-01317-y. eCollection 2024 Jun. J Diabetes Metab Disord. 2023. PMID: 38932869 Free PMC article. Review.
References
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. - PubMed
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. - PubMed
- Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem. Sci. 2007;32:189–197. - PubMed
- Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl):S8–S13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL62572/HL/NHLBI NIH HHS/United States
- R01 HL062572-12/HL/NHLBI NIH HHS/United States
- R01 HL132574/HL/NHLBI NIH HHS/United States
- HL091168/HL/NHLBI NIH HHS/United States
- C06 RR018928/RR/NCRR NIH HHS/United States
- R01 HL091168-01A1/HL/NHLBI NIH HHS/United States
- R01 HL091168/HL/NHLBI NIH HHS/United States
- R01 HL062572/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases